3 Know your bug
- Recognise that there are different ways to approach your analyses depending on which species you’re analysing.
- List the three main questions that should be asked about the organism you are working with, which will determine downstream analyses steps.
- Describe, at a high-level, the main analyses steps involved in each case.
Before starting your analysis, it’s important to understand the characteristics of the species you’re working with as this will determine how best to proceed. In particular, you need to understand how much genetic diversity or plasticity is present in your bug and whether or not it recombines. For instance the approach you take to build a phylogenetic tree of Mycobacterium tuberculosis genomes will differ from the methods you’d use to build a tree with Escherichia coli. To help with making your decision, we’ve provided a flowchart which outlines the best approach to take depending on what your dataset is composed of:
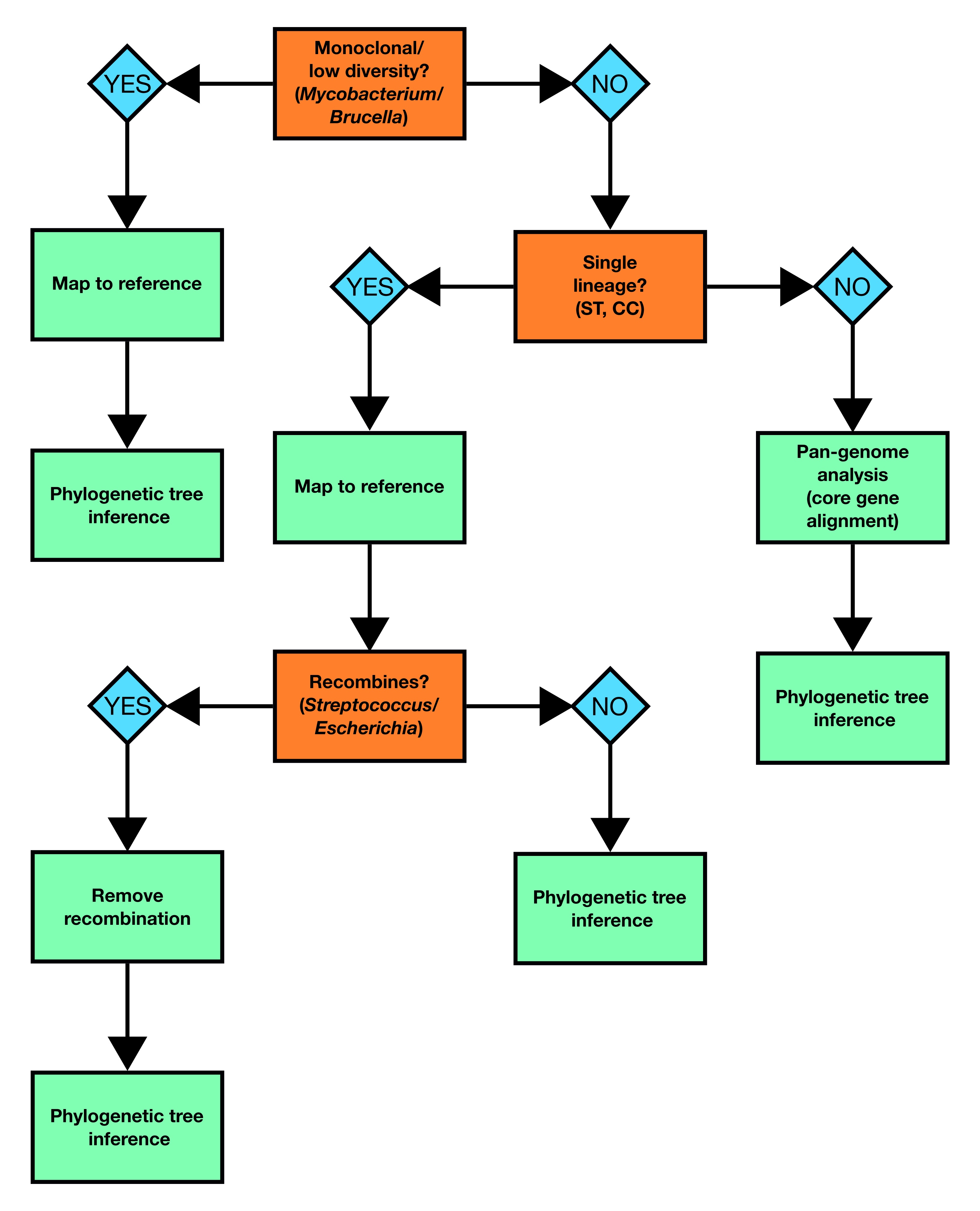
This week, we’re going to work with three different bacterial species that each require a different analysis path through the diagram above.
- Mycobacterium tuberculosis - a monoclonal (i.e. relatively little genomic diversity) organism that doesn’t recombine so we’ll build a phylogenetic tree by mapping to a reference.
- Staphylococcus aureus - a diverse organism with low levels of recombination. We’re going to be working with a dataset consisting of more than a single lineage so we’ll take the approach of building a core gene alignment to use to infer our phylogenetic tree.
- Streptococcus pneumoniae - a diverse organism that frequently recombines. As the dataset consists of isolates from a single lineage/serotype, we’re going to map to a reference and identify and remove recombinant regions before building our phylogenetic tree.
3.1 Summary
- Three main things should be considered when choosing an analysis workflow for bacterial sequencing data:
- How diverse is the species? Monoclonal species usually have lower diversity in the population.
- Do your isolates likely come from multiple lineages?
- Is bacterial recombination common in your species (transformation, transduction, conjugation)?
- Depending on the answer to these questions, your analyses workflow may involve:
- Mapping to a reference genome or using a pan-genome approach.
- Including a recombination removal step.
- The end goal of most bacterial genomics projects is the generation of a phylogenetic tree representing the relationships and diversity of your isolates.