library(tidyverse)15 Additional exercises
15.1 Section setup
# A Python data analysis and manipulation tool
import pandas as pd
# Python equivalent of `ggplot2`
from plotnine import *
# If using seaborn for plotting
import seaborn as sns
import matplotlib.pyplot as plt15.2 DA1: getting started
This is a hands-on demo.
Live demo RStudio:
- open RStudio
- highlight panels
- change default settings (
.Rdataand not save workspace) - set up new RProject
data-analysis - create the subfolders (
data,images,scripts) - create a script
Live demo JupyterLab:
- open JupyterLab
- highlight Launcher
- show how to check working directory, change if needed (
data-analysis) - create the subfolders (
data,images,scripts) - create a Notebook
15.3 DA1: data types
Important to realise that there often is some order to how data types are interpreted. For example, look at the following:
example <- c(22, 87, NA, 32)class(example)[1] "numeric"example <- c(22, 87, NA, "unsure")class(example)[1] "character"15.4 DA1: indexing
A bit of a technical detail, but an important one, particularly if you’re using both R and Python. The main focus of the example is not on the subsetting, it’s on the fact that R has 1-based indexing and Python has 0-based indexing.
Furthermore, that R’s indexing is inclusive, and Python’s exclusive (comparing 1:3 in R vs Python). That said, it’s to create awareness of the fact that indexing has to start somewhere and that it can be different between programming languages.
winnings <- c("first", "second", "third", "fourth")The positional index is as follows:
seq_along(winnings)[1] 1 2 3 4So, if we wanted to select the first and third values, we’d do:
winnings[c(1, 3)][1] "first" "third"winnings = ["first", "second", "third", "fourth"]The positional index is as follows:
list(range(len(winnings)))[0, 1, 2, 3]So, if we wanted to select the first and third values, we’d do:
winnings[0]'first'winnings[2]'third'15.5 DA2: data exploration
Read in the data:
infections <- read_csv("data/infections.csv")And have a look:
head(infections)# A tibble: 6 × 11
patient_id hospital quarter infection_type vaccination_status age_group
<chr> <chr> <chr> <chr> <chr> <chr>
1 ID_0001 hospital_3 Q2 none <NA> 65+
2 ID_0002 hospital_3 Q2 viral <NA> 18 - 64
3 ID_0003 hospital_2 Q2 none unknown 65+
4 ID_0004 hospital_2 Q3 fungal unvaccinated < 18
5 ID_0005 hospital_3 Q2 fungal vaccinated 65+
6 ID_0006 hospital_5 Q3 none vaccinated 65+
# ℹ 5 more variables: icu_admission <lgl>, symptoms_count <dbl>,
# systolic_pressure <dbl>, body_temperature <dbl>, crp_level <dbl>Read in the data:
infections = pd.read_csv("data/infections.csv")And have a look:
infections.head() patient_id hospital ... body_temperature crp_level
0 ID_0001 hospital_3 ... 37.8 12.05
1 ID_0002 hospital_3 ... 39.1 8.11
2 ID_0003 hospital_2 ... 38.5 5.24
3 ID_0004 hospital_2 ... 39.4 41.73
4 ID_0005 hospital_3 ... 36.9 10.51
[5 rows x 11 columns]15.5.1 Data structure
Number of rows & columns:
nrow(infections)[1] 1400ncol(infections)[1] 11infections.shape[0]1400infections.shape[1]11It’s good to look at the column attributes: what type of columns are we dealing with and is it what we expect?
summary(infections) patient_id hospital quarter infection_type
Length:1400 Length:1400 Length:1400 Length:1400
Class :character Class :character Class :character Class :character
Mode :character Mode :character Mode :character Mode :character
vaccination_status age_group icu_admission symptoms_count
Length:1400 Length:1400 Mode :logical Min. : 0.000
Class :character Class :character FALSE:814 1st Qu.: 6.000
Mode :character Mode :character TRUE :513 Median : 9.000
NA's :73 Mean : 8.549
3rd Qu.:11.000
Max. :21.000
NA's :67
systolic_pressure body_temperature crp_level
Min. : 87.0 Min. :36.30 Min. : 1.000
1st Qu.:118.0 1st Qu.:38.20 1st Qu.: 9.117
Median :125.0 Median :38.80 Median :16.215
Mean :125.1 Mean :38.75 Mean :19.465
3rd Qu.:132.0 3rd Qu.:39.40 3rd Qu.:26.433
Max. :163.0 Max. :41.50 Max. :58.860
NA's :74 NA's :67 NA's :156 str(infections)spc_tbl_ [1,400 × 11] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
$ patient_id : chr [1:1400] "ID_0001" "ID_0002" "ID_0003" "ID_0004" ...
$ hospital : chr [1:1400] "hospital_3" "hospital_3" "hospital_2" "hospital_2" ...
$ quarter : chr [1:1400] "Q2" "Q2" "Q2" "Q3" ...
$ infection_type : chr [1:1400] "none" "viral" "none" "fungal" ...
$ vaccination_status: chr [1:1400] NA NA "unknown" "unvaccinated" ...
$ age_group : chr [1:1400] "65+" "18 - 64" "65+" "< 18" ...
$ icu_admission : logi [1:1400] FALSE FALSE TRUE TRUE TRUE FALSE ...
$ symptoms_count : num [1:1400] 1 6 3 7 7 5 10 12 13 7 ...
$ systolic_pressure : num [1:1400] 117 115 120 129 114 124 133 120 124 127 ...
$ body_temperature : num [1:1400] 37.8 39.1 38.5 39.4 36.9 36.8 39.4 39.3 39.6 39.1 ...
$ crp_level : num [1:1400] 12.05 8.11 5.24 41.73 10.51 ...
- attr(*, "spec")=
.. cols(
.. patient_id = col_character(),
.. hospital = col_character(),
.. quarter = col_character(),
.. infection_type = col_character(),
.. vaccination_status = col_character(),
.. age_group = col_character(),
.. icu_admission = col_logical(),
.. symptoms_count = col_double(),
.. systolic_pressure = col_double(),
.. body_temperature = col_double(),
.. crp_level = col_double()
.. )
- attr(*, "problems")=<externalptr> infections.describe() symptoms_count systolic_pressure body_temperature crp_level
count 1333.000000 1326.000000 1333.000000 1244.000000
mean 8.549137 125.070136 38.751988 19.464751
std 3.825004 10.346761 0.883689 13.083907
min 0.000000 87.000000 36.300000 1.000000
25% 6.000000 118.000000 38.200000 9.117500
50% 9.000000 125.000000 38.800000 16.215000
75% 11.000000 132.000000 39.400000 26.432500
max 21.000000 163.000000 41.500000 58.86000015.5.2 Quality control checks
It’s good to do some basic sanity / quality control checks. For example, if there are different categories in a column, do all the categories we expect show up or are there missing ones / misspelled etc.?
For example, we can check the unique values in a column:
unique(infections$infection_type)[1] "none" "viral" "fungal" NA "bacterial"infections["infection_type"].unique()array(['none', 'viral', 'fungal', nan, 'bacterial'], dtype=object)We can count the number of missing values in the column infection_type.
You read the code “inside-out”:
sum(is.na(infections$infection_type))[1] 74infections["infection_type"].isna().sum()np.int64(74)15.6 DA2: subsetting tables
Let’s select patient_id:
infections$patient_idinfections.patient_id0 ID_0001
1 ID_0002
2 ID_0003
3 ID_0004
4 ID_0005
...
1395 ID_1396
1396 ID_1397
1397 ID_1398
1398 ID_1399
1399 ID_1400
Name: patient_id, Length: 1400, dtype: objectOr more than 1 column, by column name:
infections[, c("patient_id", "systolic_pressure")]# A tibble: 1,400 × 2
patient_id systolic_pressure
<chr> <dbl>
1 ID_0001 117
2 ID_0002 115
3 ID_0003 120
4 ID_0004 129
5 ID_0005 114
6 ID_0006 124
7 ID_0007 133
8 ID_0008 120
9 ID_0009 124
10 ID_0010 127
# ℹ 1,390 more rowsinfections[["patient_id", "systolic_pressure"]] patient_id systolic_pressure
0 ID_0001 117.0
1 ID_0002 115.0
2 ID_0003 120.0
3 ID_0004 129.0
4 ID_0005 114.0
... ... ...
1395 ID_1396 118.0
1396 ID_1397 117.0
1397 ID_1398 137.0
1398 ID_1399 135.0
1399 ID_1400 129.0
[1400 rows x 2 columns]Combine this with selecting only a subset of rows, let’s say the first three rows.
infections[1:3, c("patient_id", "systolic_pressure")]# A tibble: 3 × 2
patient_id systolic_pressure
<chr> <dbl>
1 ID_0001 117
2 ID_0002 115
3 ID_0003 120We need to be aware of the zero-based indexing, also noting that the value after the : is not included:
infections[["patient_id", "systolic_pressure"]].iloc[0:3] patient_id systolic_pressure
0 ID_0001 117.0
1 ID_0002 115.0
2 ID_0003 120.015.7 DA2: simple plots
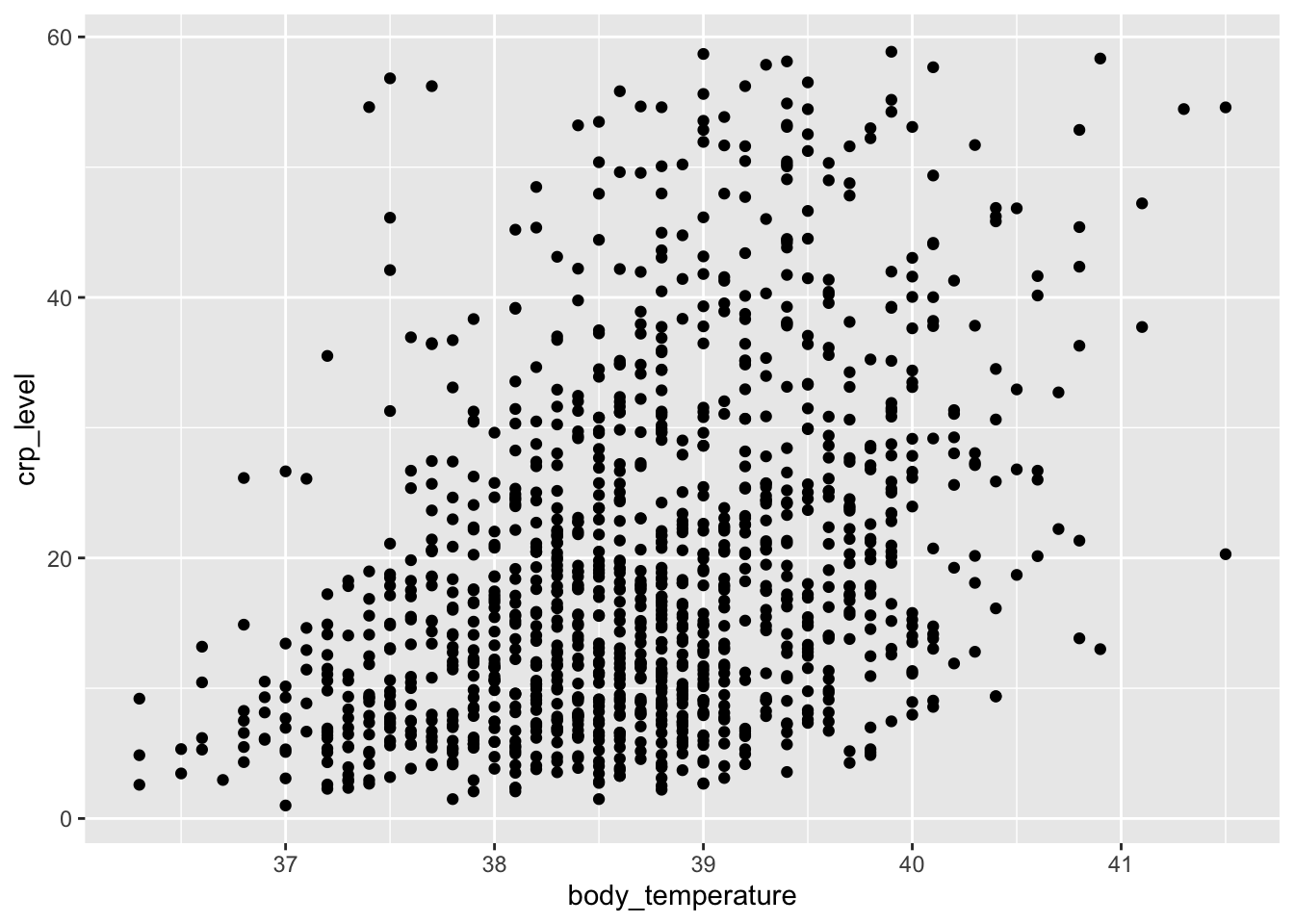
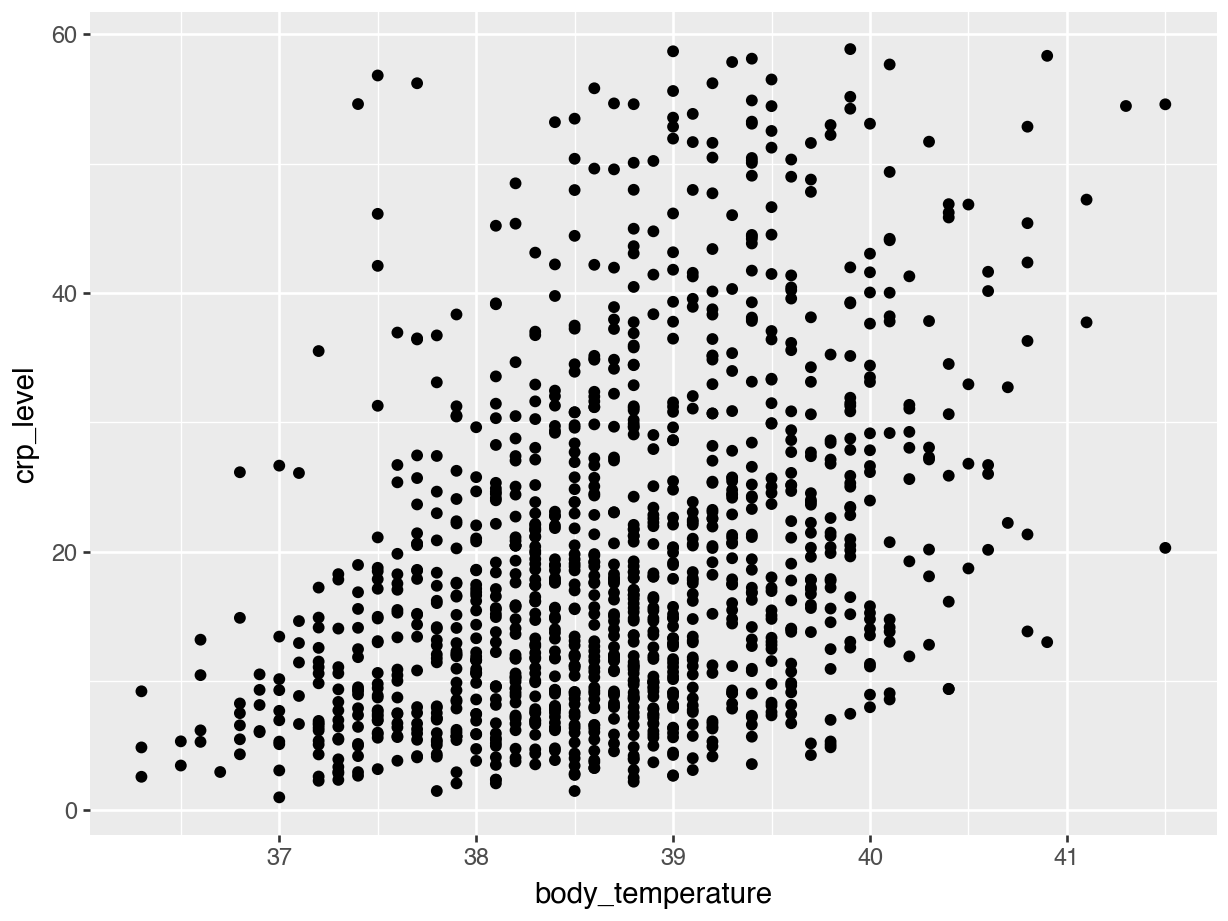
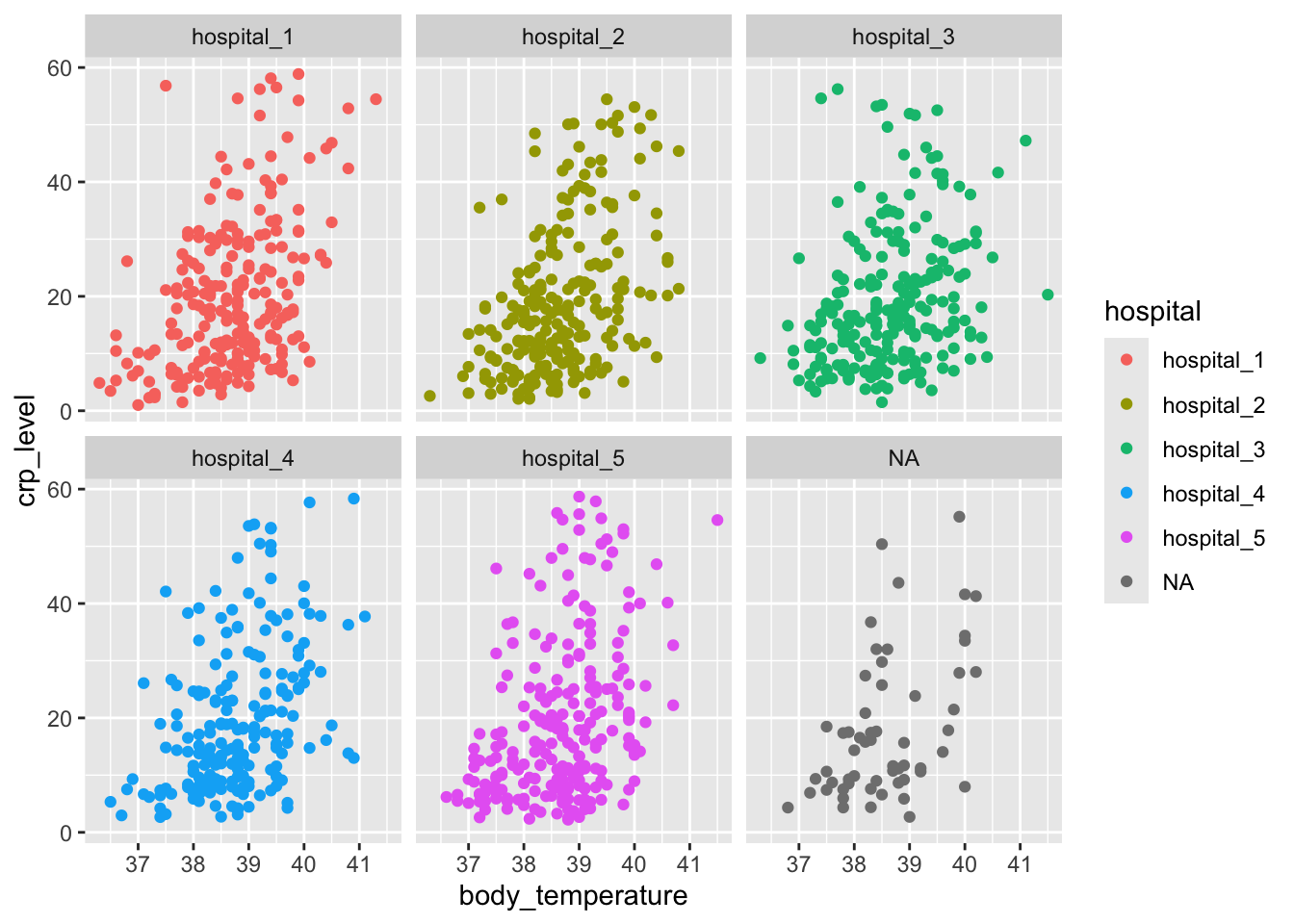
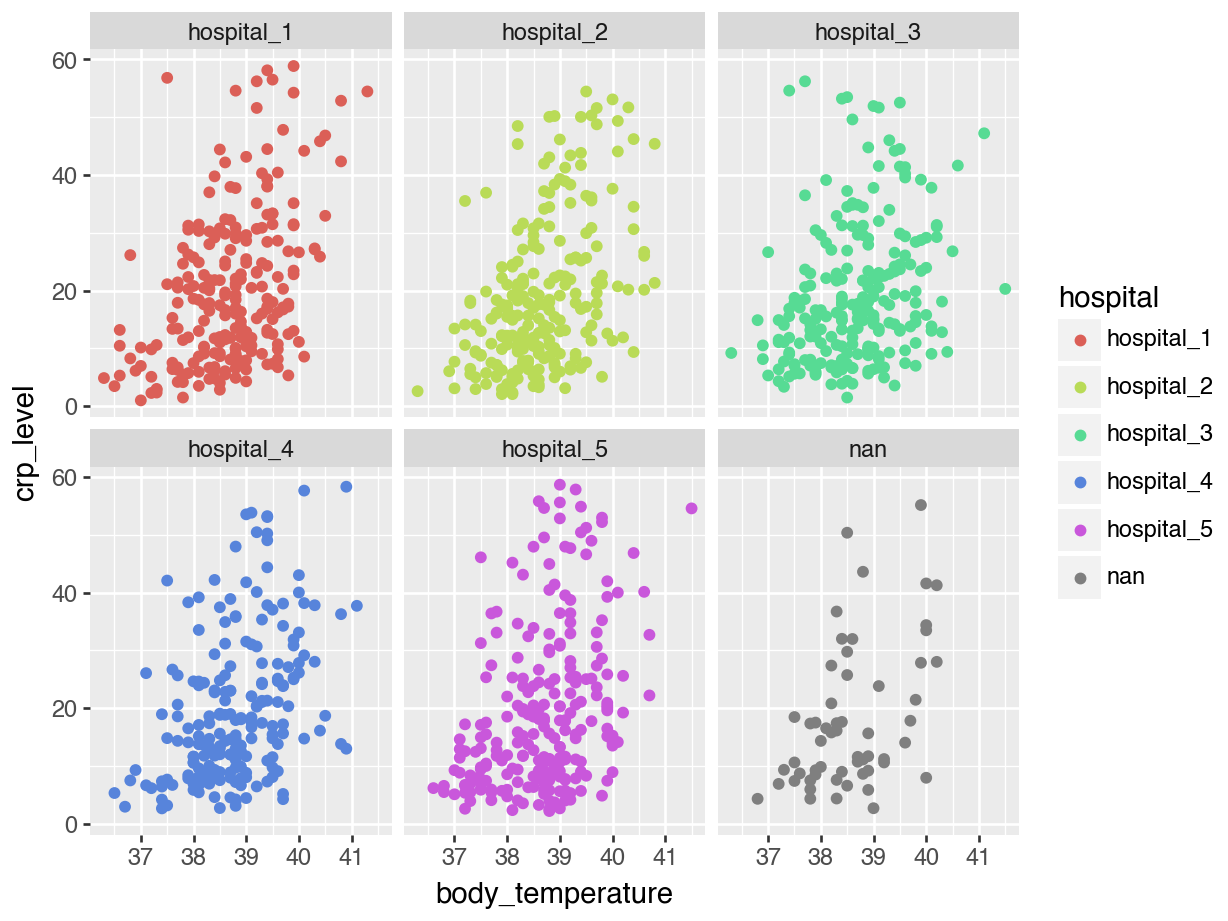
Let’s start with a simple scatterplot, where we plot body_temperature on the x-axis and crp_level on the y-axis.
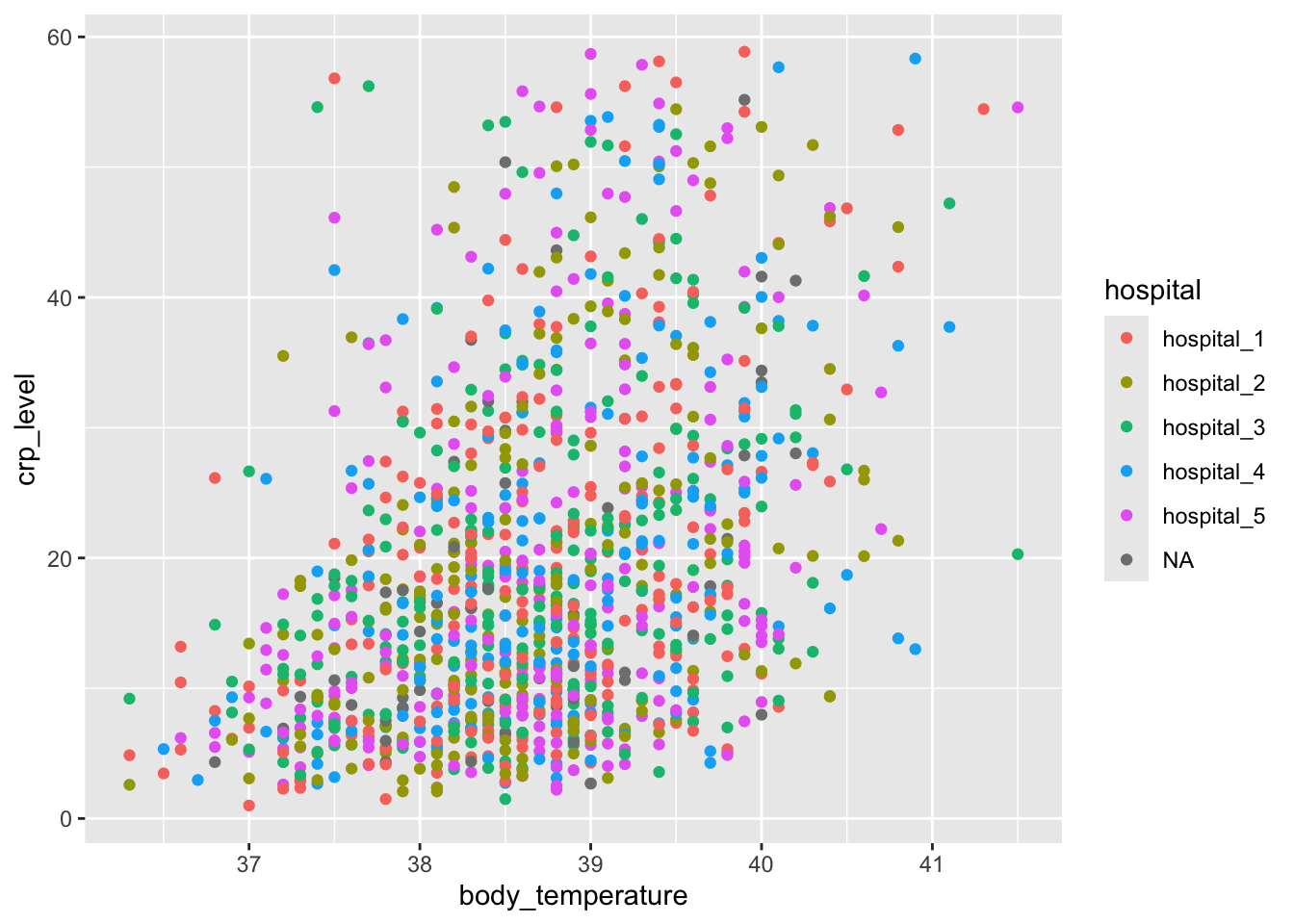
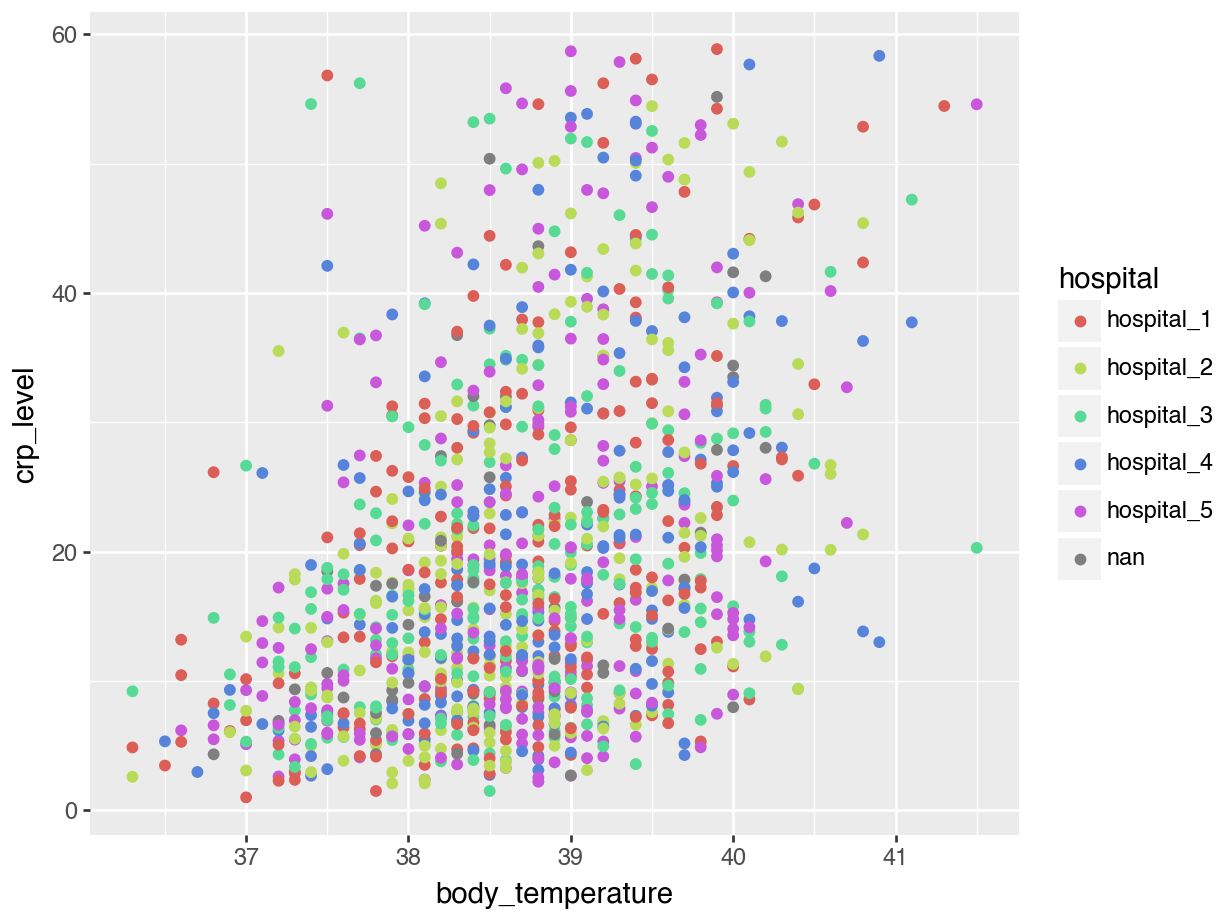
We can explore this a bit further. For example, we can colour the points based on the hospital variable, to see if there are any patterns across the different hospitals:
15.8 DA2: facetting data
When we plotted body_temperature and crp_level against each other and coloured the data based on hospital, we ended up with a rather unclear plot. This is probably because there are no clear differences between the hospitals. However, we can separate these data a bit more clearly by using facets.
ggplot(infections, aes(x = body_temperature, y = crp_level,
colour = hospital)) +
geom_point() +
facet_wrap(facets = vars(hospital))Warning: Removed 215 rows containing missing values or values outside the scale range
(`geom_point()`).15.9 DA3: selecting columns
We’re practising two things:
- selecting columns
- creating columns (and highlight the use by plotting)
Let’s see which columns we have:
colnames(infections) [1] "patient_id" "hospital" "quarter"
[4] "infection_type" "vaccination_status" "age_group"
[7] "icu_admission" "symptoms_count" "systolic_pressure"
[10] "body_temperature" "crp_level" Let’s say we only wanted to select some of these.
select(infections, patient_id, body_temperature)# A tibble: 1,400 × 2
patient_id body_temperature
<chr> <dbl>
1 ID_0001 37.8
2 ID_0002 39.1
3 ID_0003 38.5
4 ID_0004 39.4
5 ID_0005 36.9
6 ID_0006 36.8
7 ID_0007 39.4
8 ID_0008 39.3
9 ID_0009 39.6
10 ID_0010 39.1
# ℹ 1,390 more rowsOr select by data type:
select(infections, where(is.numeric))# A tibble: 1,400 × 4
symptoms_count systolic_pressure body_temperature crp_level
<dbl> <dbl> <dbl> <dbl>
1 1 117 37.8 12.0
2 6 115 39.1 8.11
3 3 120 38.5 5.24
4 7 129 39.4 41.7
5 7 114 36.9 10.5
6 5 124 36.8 6.57
7 10 133 39.4 53.1
8 12 120 39.3 NA
9 13 124 39.6 50.3
10 7 127 39.1 13.0
# ℹ 1,390 more rowsOr based on a certain phrase within the column heading:
select(infections, contains("_id"))# A tibble: 1,400 × 1
patient_id
<chr>
1 ID_0001
2 ID_0002
3 ID_0003
4 ID_0004
5 ID_0005
6 ID_0006
7 ID_0007
8 ID_0008
9 ID_0009
10 ID_0010
# ℹ 1,390 more rows15.10 DA3: creating columns
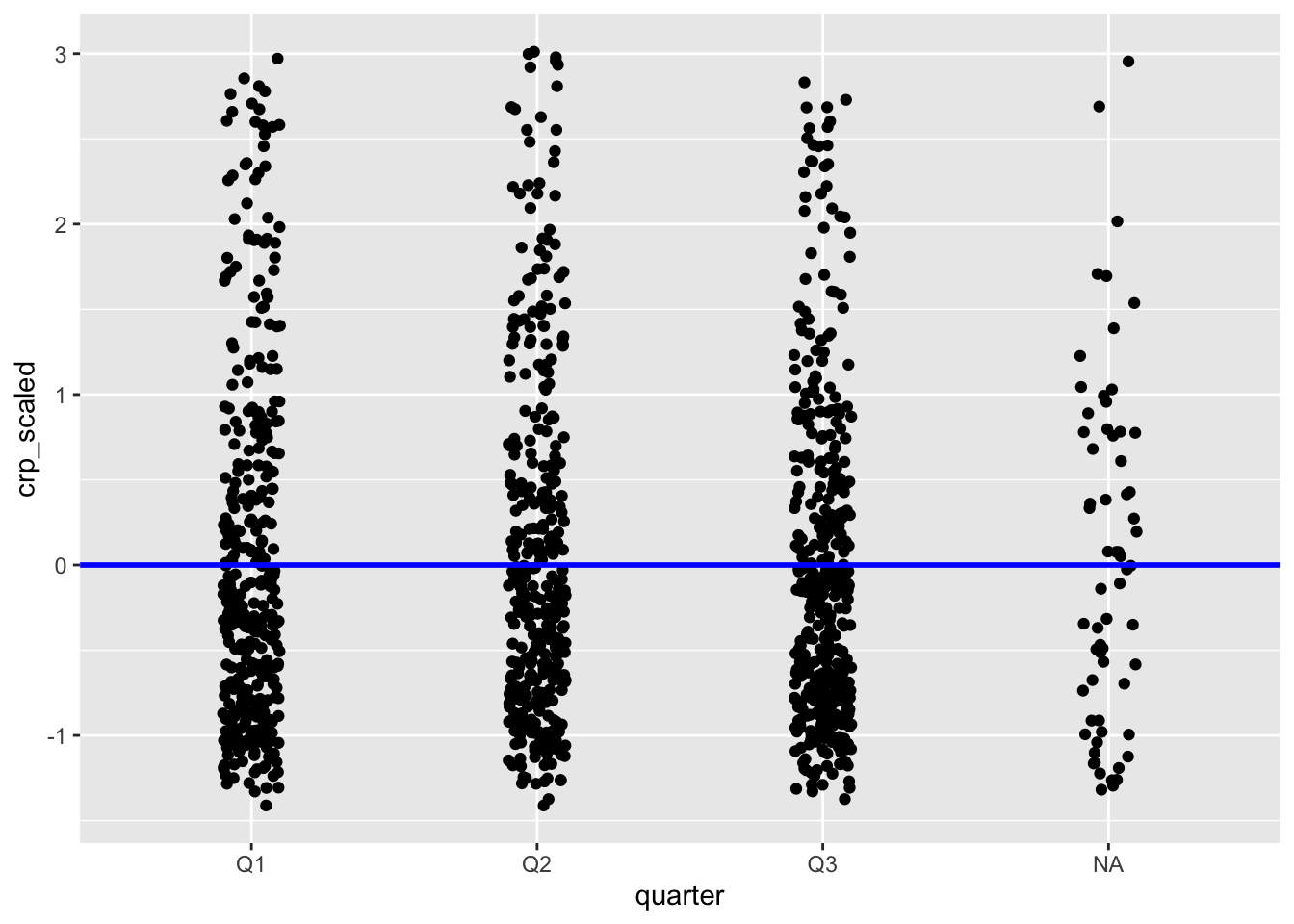
There aren’t too many variables in our data set that we can transform. However, the CRP levels are a good example. The CRP (C-reactive protein) is a continuous biomarker, often used clinically to indicate inflammation or infection severity. It can be skewed at times, so scaling it could be useful if you’re making comparisons across groups.
This falls well outside the scope of this course, but it’s possible that the question comes up. We could also log-transform our data if there is an issue with skewing etc. Very simply & briefly put:
- use
log()when the distribution shape is the issue (e.g. skew, tails) - use
scale()when the unit or range is the issue (often when comparing across different variables with different units) - using both can also be helpful, where you log-transform first, then scale the data to use in modelling.
We can scale the data where, for each value, we subtract the mean and divide by the standard deviation.
In R, we can use the scale() function to do this.
mutate(infections, crp_scaled = scale(crp_level))# A tibble: 1,400 × 12
patient_id hospital quarter infection_type vaccination_status age_group
<chr> <chr> <chr> <chr> <chr> <chr>
1 ID_0001 hospital_3 Q2 none <NA> 65+
2 ID_0002 hospital_3 Q2 viral <NA> 18 - 64
3 ID_0003 hospital_2 Q2 none unknown 65+
4 ID_0004 hospital_2 Q3 fungal unvaccinated < 18
5 ID_0005 hospital_3 Q2 fungal vaccinated 65+
6 ID_0006 hospital_5 Q3 none vaccinated 65+
7 ID_0007 hospital_4 Q1 fungal unvaccinated 18 - 64
8 ID_0008 hospital_1 Q1 <NA> unvaccinated 18 - 64
9 ID_0009 hospital_2 Q1 viral <NA> 65+
10 ID_0010 hospital_3 Q3 none unvaccinated <NA>
# ℹ 1,390 more rows
# ℹ 6 more variables: icu_admission <lgl>, symptoms_count <dbl>,
# systolic_pressure <dbl>, body_temperature <dbl>, crp_level <dbl>,
# crp_scaled <dbl[,1]>Let’s store this in a temporary object, so we can visualise it.
example <- mutate(infections, crp_scaled = scale(crp_level))We can then plot it, adding a reference line at y = 0.
15.11 DA3: creating columns (conditional)
Sometimes we need to create new columns based on certain conditions. Let’s say we wanted to flag if patients not vaccinated and have been admitted to ICU we consider them as high risk.
We want to encode this in a column risk_factor, where we label them as "high". This would then allow us to add other values, such as "low" or "medium" at a later time.
mutate(infections,
risk_factor = ifelse(vaccination_status == "unvaccinated" & icu_admission == TRUE, "high", NA))# A tibble: 1,400 × 12
patient_id hospital quarter infection_type vaccination_status age_group
<chr> <chr> <chr> <chr> <chr> <chr>
1 ID_0001 hospital_3 Q2 none <NA> 65+
2 ID_0002 hospital_3 Q2 viral <NA> 18 - 64
3 ID_0003 hospital_2 Q2 none unknown 65+
4 ID_0004 hospital_2 Q3 fungal unvaccinated < 18
5 ID_0005 hospital_3 Q2 fungal vaccinated 65+
6 ID_0006 hospital_5 Q3 none vaccinated 65+
7 ID_0007 hospital_4 Q1 fungal unvaccinated 18 - 64
8 ID_0008 hospital_1 Q1 <NA> unvaccinated 18 - 64
9 ID_0009 hospital_2 Q1 viral <NA> 65+
10 ID_0010 hospital_3 Q3 none unvaccinated <NA>
# ℹ 1,390 more rows
# ℹ 6 more variables: icu_admission <lgl>, symptoms_count <dbl>,
# systolic_pressure <dbl>, body_temperature <dbl>, crp_level <dbl>,
# risk_factor <chr>15.12 DA4: reshaping data
To illustrate reshaping data, we’re going to simplify our data set a little bit. First, we’ll calculate the average CRP levels for each hospital, quarter, infection type, vaccination status and age group.
We’ll use that summary table to highlight some use cases for reshaping data from a long to a wide format - and back.
15.12.1 Creating a summary table for CRP
infections_summary <- infections |>
drop_na() |>
group_by(hospital, quarter, infection_type, vaccination_status, age_group) |>
summarise(mean_crp = round(mean(crp_level, na.rm = TRUE), 2)) |>
ungroup()`summarise()` has grouped output by 'hospital', 'quarter', 'infection_type',
'vaccination_status'. You can override using the `.groups` argument.Let’s look at the output. We now have 6 columns, one for each of our grouping variables and at the end the mean_crp column containing the average CRP levels (rounded to two digits).
head(infections_summary)# A tibble: 6 × 6
hospital quarter infection_type vaccination_status age_group mean_crp
<chr> <chr> <chr> <chr> <chr> <dbl>
1 hospital_1 Q1 bacterial unknown 65+ 23.7
2 hospital_1 Q1 bacterial unknown < 18 25.8
3 hospital_1 Q1 bacterial unvaccinated 18 - 64 8.35
4 hospital_1 Q1 bacterial unvaccinated 65+ 23.2
5 hospital_1 Q1 bacterial unvaccinated < 18 26.8
6 hospital_1 Q1 bacterial vaccinated 18 - 64 22.4 15.12.2 Wide table by quarter
Let’s say we’d be interested how the (average) CRP levels change over time across the different groupings. We’d be interested in having the mean_crp values for each quarter next to each other. That way we’d be able to plot quarterly average CRP levels against each other.
We can do this as follows:
infections_wide <- infections_summary |>
pivot_wider(names_from = "quarter",
values_from = "mean_crp")head(infections_wide)# A tibble: 6 × 7
hospital infection_type vaccination_status age_group Q1 Q2 Q3
<chr> <chr> <chr> <chr> <dbl> <dbl> <dbl>
1 hospital_1 bacterial unknown 65+ 23.7 NA 26.8
2 hospital_1 bacterial unknown < 18 25.8 54.5 NA
3 hospital_1 bacterial unvaccinated 18 - 64 8.35 NA NA
4 hospital_1 bacterial unvaccinated 65+ 23.2 28.6 NA
5 hospital_1 bacterial unvaccinated < 18 26.8 25.0 17.1
6 hospital_1 bacterial vaccinated 18 - 64 22.4 12.7 NA This then allows us to plot the data, for example:
15.12.3 Back to long format
We can revert back to a long format with the following:
pivot_longer(infections_wide,
cols = Q1:Q3, # columns to pivot
names_to = "quarter", # column for headings
values_to = "mean_crp") # column for values# A tibble: 534 × 6
hospital infection_type vaccination_status age_group quarter mean_crp
<chr> <chr> <chr> <chr> <chr> <dbl>
1 hospital_1 bacterial unknown 65+ Q1 23.7
2 hospital_1 bacterial unknown 65+ Q2 NA
3 hospital_1 bacterial unknown 65+ Q3 26.8
4 hospital_1 bacterial unknown < 18 Q1 25.8
5 hospital_1 bacterial unknown < 18 Q2 54.5
6 hospital_1 bacterial unknown < 18 Q3 NA
7 hospital_1 bacterial unvaccinated 18 - 64 Q1 8.35
8 hospital_1 bacterial unvaccinated 18 - 64 Q2 NA
9 hospital_1 bacterial unvaccinated 18 - 64 Q3 NA
10 hospital_1 bacterial unvaccinated 65+ Q1 23.2
# ℹ 524 more rowsAnd all is well with the world again.
15.13 DA4: joining data
In the infections data set we have a variable called hospital. This contains the following unique entries:
infections |> count(hospital)# A tibble: 6 × 2
hospital n
<chr> <int>
1 hospital_1 277
2 hospital_2 259
3 hospital_3 280
4 hospital_4 245
5 hospital_5 271
6 <NA> 68Note that for some of the hospital entries there are missing data. This is relevant later on, when we’re joining.
We’re now going to add information on the hospitals, which are stored in hospital_info.csv. Let’s read in the data:
hospital_info <- read_csv("data/hospital_info.csv")Look at the data. There are 6 distinct hospital entries, with additional information for each hospital. We have the following variables:
| Variable | Type | Description |
|---|---|---|
hospital |
character (id) | Unique hospital identifier (hospital_1 … hospital_6). |
hospital_name |
character | Official hospital name (e.g., Royal London Hospital). |
location |
character | City where the hospital is located (e.g., London, Manchester). |
bed_capacity |
integer | Approximate number of inpatient beds available at the hospital. |
teaching_hospital |
logical | Indicates if the hospital is a teaching hospital (TRUE / FALSE). |
15.13.1 Left join
First, we’ll add the hospital_info data to the infections data set. We do this with a left-join. We expect hospital info data to be added to our main infections table, if the hospital value in infections matches with the one in hospital_info.
infections_left <- left_join(infections, hospital_info, by = "hospital")Let’s just select the ID column from infections, together with all the columns that have been added.
infections_left |>
select(patient_id, hospital, hospital_name, location, bed_capacity, teaching_hospital) |>
head()# A tibble: 6 × 6
patient_id hospital hospital_name location bed_capacity teaching_hospital
<chr> <chr> <chr> <chr> <dbl> <lgl>
1 ID_0001 hospital_3 Bristol Royal I… Bristol 768 TRUE
2 ID_0002 hospital_3 Bristol Royal I… Bristol 768 TRUE
3 ID_0003 hospital_2 Manchester Gene… Manches… 849 FALSE
4 ID_0004 hospital_2 Manchester Gene… Manches… 849 FALSE
5 ID_0005 hospital_3 Bristol Royal I… Bristol 768 TRUE
6 ID_0006 hospital_5 Cardiff Univers… Cardiff 582 TRUE We can see that the hospital information is now added to the data. We don’t expect any data to have dropped, so the number of observations/rows in infections_left should match the original 1400 from infections.
nrow(infections_left)[1] 140015.13.2 Right join
So, how does a right join then differ from a left join? Well, here we’d be adding the infections data to the hospital_info data (so, in the other direction). That means that for each hospital value that exists in hospital_info it will try and find the values that match the hospital column in infections.
infections_right <- right_join(infections, hospital_info, by = "hospital")Again, the resulting table contains the data from both infections and hospital_info:
head(infections_right)# A tibble: 6 × 15
patient_id hospital quarter infection_type vaccination_status age_group
<chr> <chr> <chr> <chr> <chr> <chr>
1 ID_0001 hospital_3 Q2 none <NA> 65+
2 ID_0002 hospital_3 Q2 viral <NA> 18 - 64
3 ID_0003 hospital_2 Q2 none unknown 65+
4 ID_0004 hospital_2 Q3 fungal unvaccinated < 18
5 ID_0005 hospital_3 Q2 fungal vaccinated 65+
6 ID_0006 hospital_5 Q3 none vaccinated 65+
# ℹ 9 more variables: icu_admission <lgl>, symptoms_count <dbl>,
# systolic_pressure <dbl>, body_temperature <dbl>, crp_level <dbl>,
# hospital_name <chr>, location <chr>, bed_capacity <dbl>,
# teaching_hospital <lgl>How many observations do we expect? Let’s have a look at how many we’ve got.
nrow(infections_right)[1] 1333Here we see that we have fewer than the original 1400 observations, because we are joining in the other direction. Remember, there are missing values in our infections data set (68 in total). When we’re joining infections to hospital_info, these entries get dropped, because there is no missing value entry in the hospital column of hospital_info.
For the eagle-eyed amongst you: we have 1333 observations/rows in infections_right, whereas there are 68 missing values. We might have expected there to be 1332 rows (the difference between the number of rows in infections and the number of missing values), but there are 1333.
Why? Well, there is one entry in hospital_info that does not appear in infections: hospital_6. When we right join, this value gets retained, so this adds one additional row to the final output.
infections_right |> count(hospital)# A tibble: 6 × 2
hospital n
<chr> <int>
1 hospital_1 277
2 hospital_2 259
3 hospital_3 280
4 hospital_4 245
5 hospital_5 271
6 hospital_6 1If we wouldn’t want that, we’d use inner join.
15.13.3 Inner join
Inner join will join two tables and only retain values (based on the joining ID) that exist in both tables.
infections_inner <- inner_join(infections, hospital_info, by = "hospital")nrow(infections_inner)[1] 1332Only the 5 hospitals that appear in the infections data set are retained:
infections_inner |> count(hospital)# A tibble: 5 × 2
hospital n
<chr> <int>
1 hospital_1 277
2 hospital_2 259
3 hospital_3 280
4 hospital_4 245
5 hospital_5 271This is entirely expected, since we don’t have missing data in hospital_info (so all the missing data from infections are dropped), nor do we have a hospital_6 entry in infections (so that is dropped too).
15.13.4 Full join
If we wanted to retain all observations, regardless of which table they’re from, we’d use a full join.
infections_full <- full_join(infections, hospital_info, by = "hospital")How many rows do we expect? Well, the 1400 from infections plus the one extra (hospital_6 entry) from hospital_info:
nrow(infections_full)[1] 1401We can check that we’ve retained all values, by counting the number of observations for each hospital value:
infections_full |> count(hospital)# A tibble: 7 × 2
hospital n
<chr> <int>
1 hospital_1 277
2 hospital_2 259
3 hospital_3 280
4 hospital_4 245
5 hospital_5 271
6 hospital_6 1
7 <NA> 68We see that all hospital values are retained, including the missing values and the one hospital_6 value coming from the hospital_info data set. Success!