# A collection of R packages designed for data science
library(tidyverse)
surveys <- read_csv("data/surveys.csv")8 Manipulating columns
- Learn how to select and/or rename specific columns from a data frame.
- Be able to create new columns and modify existing ones.
8.1 Context
In the tabular data section we learned to deal with, well, tabular data in the form of our surveys data set. This data set isn’t huge, but sometimes we have many variables and we might only want to work with a subset of them. Or, we might want to create new columns based on existing data. In this section we’ll cover how we can do this.
8.2 Section setup
We’ll continue this section with the script named 03_session. If needed, add the following code to the top of your script and run it.
We’ll continue this section with the Notebook named 03_session. Add the following code to the first cell and run it.
# A Python data analysis and manipulation tool
import pandas as pd
# Python equivalent of `ggplot2`
from plotnine import *
surveys = pd.read_csv("data/surveys.csv")8.3 Selecting columns
Let’s remind ourselves of which columns we have in our surveys data set. After that, we’ll start making some changes.
colnames(surveys)[1] "record_id" "month" "day" "year"
[5] "plot_id" "species_id" "sex" "hindfoot_length"
[9] "weight" surveys.columnsIndex(['record_id', 'month', 'day', 'year', 'plot_id', 'species_id', 'sex',
'hindfoot_length', 'weight'],
dtype='object')8.3.1 Selecting individual columns
Let’s say we wanted to select only the record_id and year columns. We’ve briefly done this when we looked at subsetting rows and columns. This used some basic subsetting, using the [ ] notation.
There is an alternative way of doing this using the dplyr package - which is part of tidyverse.
We can use the select() function. This function selects columns.
select(surveys, record_id, year)# A tibble: 35,549 × 2
record_id year
<dbl> <dbl>
1 1 1977
2 2 1977
3 3 1977
4 4 1977
5 5 1977
6 6 1977
7 7 1977
8 8 1977
9 9 1977
10 10 1977
# ℹ 35,539 more rowsUsing the base R syntax, this is equivalent to surveys[, c("record_id", "year")]. Notice that with the select() function (and generally with dplyr functions) we didn’t need to quote ” the column names. This is because the first input to the function is the table name, and so everything after is assumed to be column names of that table.
If we wanted to get the resulting output in a separate tab, so we could view it better, we could wrap this inside the View() function:
View(select(surveys, record_id, year))The way we need to specify this is by giving a list of column names ["record_id", "year"] and subsetting the surveys data set with this.
The way we subset is with surveys[ ], so we end up with double square brackets: one set for the subsetting and one set for the list.
surveys[["record_id", "year"]] record_id year
0 1 1977
1 2 1977
2 3 1977
3 4 1977
4 5 1977
... ... ...
35544 35545 2002
35545 35546 2002
35546 35547 2002
35547 35548 2002
35548 35549 2002
[35549 rows x 2 columns]8.3.2 Selecting with helper functions
The select() function becomes particularly useful when we combine it with other helper functions. For example, this code will select all the columns where the column name contains the string (text) "_id":
# returns all columns where the column name contains the text "_id"
select(surveys, contains("_id"))# A tibble: 35,549 × 3
record_id plot_id species_id
<dbl> <dbl> <chr>
1 1 2 NL
2 2 3 NL
3 3 2 DM
4 4 7 DM
5 5 3 DM
6 6 1 PF
7 7 2 PE
8 8 1 DM
9 9 1 DM
10 10 6 PF
# ℹ 35,539 more rowsAnother thing we often want to do is select columns by their data type. For example, if we wanted to select all numerical columns we could do this:
select(surveys, where(is.numeric))# A tibble: 35,549 × 7
record_id month day year plot_id hindfoot_length weight
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 7 16 1977 2 32 NA
2 2 7 16 1977 3 33 NA
3 3 7 16 1977 2 37 NA
4 4 7 16 1977 7 36 NA
5 5 7 16 1977 3 35 NA
6 6 7 16 1977 1 14 NA
7 7 7 16 1977 2 NA NA
8 8 7 16 1977 1 37 NA
9 9 7 16 1977 1 34 NA
10 10 7 16 1977 6 20 NA
# ℹ 35,539 more rowsThe subsetting becomes a bit tedious when we’re looking for patterns in the column names. Here, we can instead use the .filter method of the surveys data set, and look for a string (text) where the column name contains "_id".
surveys.filter(like = "_id") record_id plot_id species_id
0 1 2 NL
1 2 3 NL
2 3 2 DM
3 4 7 DM
4 5 3 DM
... ... ... ...
35544 35545 15 AH
35545 35546 15 AH
35546 35547 10 RM
35547 35548 7 DO
35548 35549 5 NaN
[35549 rows x 3 columns]Another thing we often want to do is select columns by their data type. For example, if we wanted to select all numerical columns we could do this:
surveys.select_dtypes(include = "number") record_id month day year plot_id hindfoot_length weight
0 1 7 16 1977 2 32.0 NaN
1 2 7 16 1977 3 33.0 NaN
2 3 7 16 1977 2 37.0 NaN
3 4 7 16 1977 7 36.0 NaN
4 5 7 16 1977 3 35.0 NaN
... ... ... ... ... ... ... ...
35544 35545 12 31 2002 15 NaN NaN
35545 35546 12 31 2002 15 NaN NaN
35546 35547 12 31 2002 10 15.0 14.0
35547 35548 12 31 2002 7 36.0 51.0
35548 35549 12 31 2002 5 NaN NaN
[35549 rows x 7 columns]8.3.3 Selecting a range of columns
Let’s say we’re interested in all the columns from record_id to year.
In that case, we can use the : symbol.
# returns all columns between and including record_id and year
select(surveys, record_id:year)# A tibble: 35,549 × 4
record_id month day year
<dbl> <dbl> <dbl> <dbl>
1 1 7 16 1977
2 2 7 16 1977
3 3 7 16 1977
4 4 7 16 1977
5 5 7 16 1977
6 6 7 16 1977
7 7 7 16 1977
8 8 7 16 1977
9 9 7 16 1977
10 10 7 16 1977
# ℹ 35,539 more rowsWe can also combine this with the previous method:
# returns all columns between and including record_id and year
# and all columns where the column name contains the text "_id"
select(surveys, record_id:year, contains("_id"))# A tibble: 35,549 × 6
record_id month day year plot_id species_id
<dbl> <dbl> <dbl> <dbl> <dbl> <chr>
1 1 7 16 1977 2 NL
2 2 7 16 1977 3 NL
3 3 7 16 1977 2 DM
4 4 7 16 1977 7 DM
5 5 7 16 1977 3 DM
6 6 7 16 1977 1 PF
7 7 7 16 1977 2 PE
8 8 7 16 1977 1 DM
9 9 7 16 1977 1 DM
10 10 7 16 1977 6 PF
# ℹ 35,539 more rowsIn that case, we can use the : symbol, in combination with the .loc indexer.
surveys.loc[:, "record_id":"year"] record_id month day year
0 1 7 16 1977
1 2 7 16 1977
2 3 7 16 1977
3 4 7 16 1977
4 5 7 16 1977
... ... ... ... ...
35544 35545 12 31 2002
35545 35546 12 31 2002
35546 35547 12 31 2002
35547 35548 12 31 2002
35548 35549 12 31 2002
[35549 rows x 4 columns]8.3.4 Unselecting columns
Lastly, we can also unselect columns. This can be useful when you want most columns, apart from some.
To do this, we use the - symbol before the column name.
# returns all columns apart from record_id
select(surveys, -record_id)# A tibble: 35,549 × 8
month day year plot_id species_id sex hindfoot_length weight
<dbl> <dbl> <dbl> <dbl> <chr> <chr> <dbl> <dbl>
1 7 16 1977 2 NL M 32 NA
2 7 16 1977 3 NL M 33 NA
3 7 16 1977 2 DM F 37 NA
4 7 16 1977 7 DM M 36 NA
5 7 16 1977 3 DM M 35 NA
6 7 16 1977 1 PF M 14 NA
7 7 16 1977 2 PE F NA NA
8 7 16 1977 1 DM M 37 NA
9 7 16 1977 1 DM F 34 NA
10 7 16 1977 6 PF F 20 NA
# ℹ 35,539 more rowsTo do this, we use the .drop attribute. Here, we only unselect one column, but we can easily extend this by providing a list of columns do the column = argument.
surveys.drop(columns = "record_id") month day year plot_id species_id sex hindfoot_length weight
0 7 16 1977 2 NL M 32.0 NaN
1 7 16 1977 3 NL M 33.0 NaN
2 7 16 1977 2 DM F 37.0 NaN
3 7 16 1977 7 DM M 36.0 NaN
4 7 16 1977 3 DM M 35.0 NaN
... ... ... ... ... ... ... ... ...
35544 12 31 2002 15 AH NaN NaN NaN
35545 12 31 2002 15 AH NaN NaN NaN
35546 12 31 2002 10 RM F 15.0 14.0
35547 12 31 2002 7 DO M 36.0 51.0
35548 12 31 2002 5 NaN NaN NaN NaN
[35549 rows x 8 columns]8.4 Renaming and reshuffling columns
8.4.1 Renaming columns
For example, we might want to change the weight column name to weight_g, to reflect that the values are in grams.
We can use the rename() function to change a column name. We do this as follows:
rename(surveys, weight_g = weight)# A tibble: 35,549 × 9
record_id month day year plot_id species_id sex hindfoot_length weight_g
<dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <dbl> <dbl>
1 1 7 16 1977 2 NL M 32 NA
2 2 7 16 1977 3 NL M 33 NA
3 3 7 16 1977 2 DM F 37 NA
4 4 7 16 1977 7 DM M 36 NA
5 5 7 16 1977 3 DM M 35 NA
6 6 7 16 1977 1 PF M 14 NA
7 7 7 16 1977 2 PE F NA NA
8 8 7 16 1977 1 DM M 37 NA
9 9 7 16 1977 1 DM F 34 NA
10 10 7 16 1977 6 PF F 20 NA
# ℹ 35,539 more rowsWe can use the .rename() method of the surveys DataFrame:
surveys.rename(columns = {'weight': 'weight_g'}) record_id month day year ... species_id sex hindfoot_length weight_g
0 1 7 16 1977 ... NL M 32.0 NaN
1 2 7 16 1977 ... NL M 33.0 NaN
2 3 7 16 1977 ... DM F 37.0 NaN
3 4 7 16 1977 ... DM M 36.0 NaN
4 5 7 16 1977 ... DM M 35.0 NaN
... ... ... ... ... ... ... ... ... ...
35544 35545 12 31 2002 ... AH NaN NaN NaN
35545 35546 12 31 2002 ... AH NaN NaN NaN
35546 35547 12 31 2002 ... RM F 15.0 14.0
35547 35548 12 31 2002 ... DO M 36.0 51.0
35548 35549 12 31 2002 ... NaN NaN NaN NaN
[35549 rows x 9 columns]8.4.2 Reshuffling columns
It might be that you want to reorder/reshuffle a column. Here, the year column is our fourth variable. Let’s say we’d want to move this to the second position (after record_id).
We can use the relocate() function to do this. The function has several arguments, starting with ., such as .before = or .after =. These allow you to specify where you want to reinsert the column.
relocate(surveys, year, .after = record_id)# A tibble: 35,549 × 9
record_id year month day plot_id species_id sex hindfoot_length weight
<dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <dbl> <dbl>
1 1 1977 7 16 2 NL M 32 NA
2 2 1977 7 16 3 NL M 33 NA
3 3 1977 7 16 2 DM F 37 NA
4 4 1977 7 16 7 DM M 36 NA
5 5 1977 7 16 3 DM M 35 NA
6 6 1977 7 16 1 PF M 14 NA
7 7 1977 7 16 2 PE F NA NA
8 8 1977 7 16 1 DM M 37 NA
9 9 1977 7 16 1 DM F 34 NA
10 10 1977 7 16 6 PF F 20 NA
# ℹ 35,539 more rowsUnlike in R, there isn’t a very clear, straightforward way of reinserting columns in a pandas DataFrame. We could show you convoluted ways of doing so, but at this point that’s just confusing. So, we’ll leave you with a link to a Stackoverflow solution.
8.5 Creating new columns
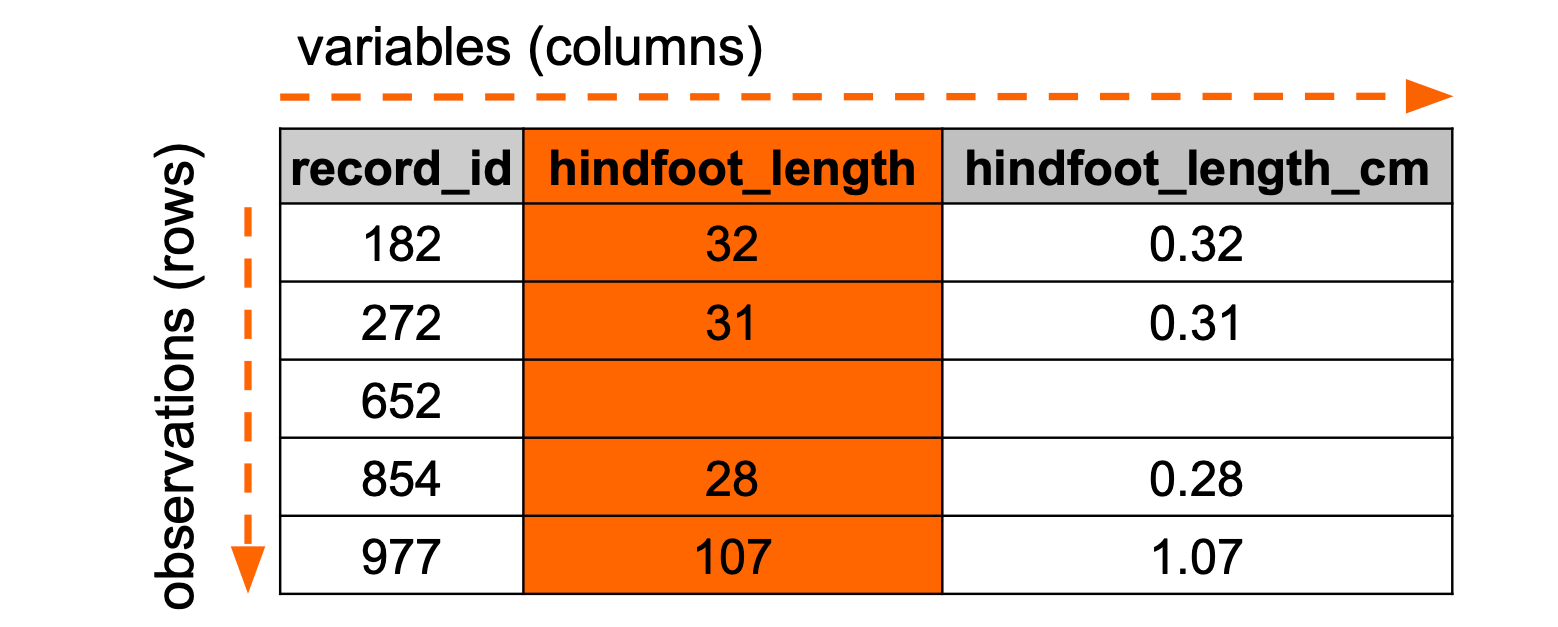
Sometimes we need to create new columns. For example, we might have a variable that is not in the unit of measurement we need (e.g. in millimeters, instead of centimeters).
Conceptually, that looks something like this:
Let’s illustrate this with an example on our surveys data. Let’s say we wanted to get hindfoot_length in centimeters, instead of millimeters. We’d have to go through each row, take the hindfoot_length value and divide it by 10. We then need to store this output in a column called, for example, hindfoot_length_cm.
We can use the mutate() function to create new columns:
mutate(surveys, hindfoot_length_cm = hindfoot_length / 10)# A tibble: 35,549 × 10
record_id month day year plot_id species_id sex hindfoot_length weight
<dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <dbl> <dbl>
1 1 7 16 1977 2 NL M 32 NA
2 2 7 16 1977 3 NL M 33 NA
3 3 7 16 1977 2 DM F 37 NA
4 4 7 16 1977 7 DM M 36 NA
5 5 7 16 1977 3 DM M 35 NA
6 6 7 16 1977 1 PF M 14 NA
7 7 7 16 1977 2 PE F NA NA
8 8 7 16 1977 1 DM M 37 NA
9 9 7 16 1977 1 DM F 34 NA
10 10 7 16 1977 6 PF F 20 NA
# ℹ 35,539 more rows
# ℹ 1 more variable: hindfoot_length_cm <dbl>We use the square brackets to define the name of the new column, then specify what needs to go in the new column:
surveys["hindfoot_length_cm"] = surveys["hindfoot_length"] / 10Although it has created the column, we can’t quite see it because we have too many columns. So, let’s save the new column to the data set and then select the relevant columns.
First, we update our data:
surveys <- mutate(surveys, hindfoot_length_cm = hindfoot_length / 10)Next, we can select the columns.
select(surveys, record_id, hindfoot_length, hindfoot_length_cm)# A tibble: 35,549 × 3
record_id hindfoot_length hindfoot_length_cm
<dbl> <dbl> <dbl>
1 1 32 3.2
2 2 33 3.3
3 3 37 3.7
4 4 36 3.6
5 5 35 3.5
6 6 14 1.4
7 7 NA NA
8 8 37 3.7
9 9 34 3.4
10 10 20 2
# ℹ 35,539 more rowsOur previous step already added the new column to the DataFrame, so we can directly select the relevant columns, by giving a list of the columns we’re interested in:
surveys[['record_id', 'hindfoot_length', 'hindfoot_length_cm']] record_id hindfoot_length hindfoot_length_cm
0 1 32.0 3.2
1 2 33.0 3.3
2 3 37.0 3.7
3 4 36.0 3.6
4 5 35.0 3.5
... ... ... ...
35544 35545 NaN NaN
35545 35546 NaN NaN
35546 35547 15.0 1.5
35547 35548 36.0 3.6
35548 35549 NaN NaN
[35549 rows x 3 columns]We can see that each value in hindfoot_length_cm is a tenth of the value of hindfoot_length. This is exactly what we expected!
8.6 Exercises
8.6.1 Selecting columns: infections
8.6.2 Creating columns: body_temperature_f
8.6.3 Creating and plotting columns
8.7 Summary
- We have several functions available that allow us to select, move, rename and create new columns