B Solutions - Dimensionality reduction
Solutions to exercises of dimensionality reduction chapter.
B.1 Exercise 1.
Read in the corresponding spreadsheet into the R environment as a data frame variable.
library(tidyverse)
set.seed(12345)
sc_rna <- read_csv(file = "data/PGC_transcriptomics/PGC_transcriptomics.csv")
metadata <- sc_rna %>%
slice( 1:4) %>%
pivot_longer( cols=-Sample, names_to = 'cell_type', values_to = 'index') %>%
pivot_wider( names_from = Sample, values_from = index) %>%
mutate(group=str_remove(cell_type, '\\..*$')) %>%
mutate_if( is.numeric, as.factor)
sc_rna_fil <- sc_rna %>%
slice(-c(1:4)) %>%
column_to_rownames(var='Sample') %>%
as.matrix()
genenames <- rownames(sc_rna_fil)
cell_type <- colnames(sc_rna_fil) We can run tSNE using the following command:
library(Rtsne)
set.seed(1)
tsne_model_1 = Rtsne(as.matrix(t(sc_rna_fil)), check_duplicates=FALSE, pca=TRUE, perplexity=100, theta=0.5, dims=2)As we did previously, we can plot the results using:
sc_2d_data <- tsne_model_1$Y %>%
as.data.frame() %>%
rename( x=V1,y=V2) %>%
mutate(cell_type = cell_type ) %>%
mutate( cell_group = str_remove(cell_type, '\\..*$'))
ggplot(data=sc_2d_data) +
geom_point( mapping=aes(x=x,y=y, color=cell_group), alpha=0.5) +
scale_x_continuous(limits = c(-20,20)) +
scale_y_continuous(limits = c(-20,20)) +
theme_classic()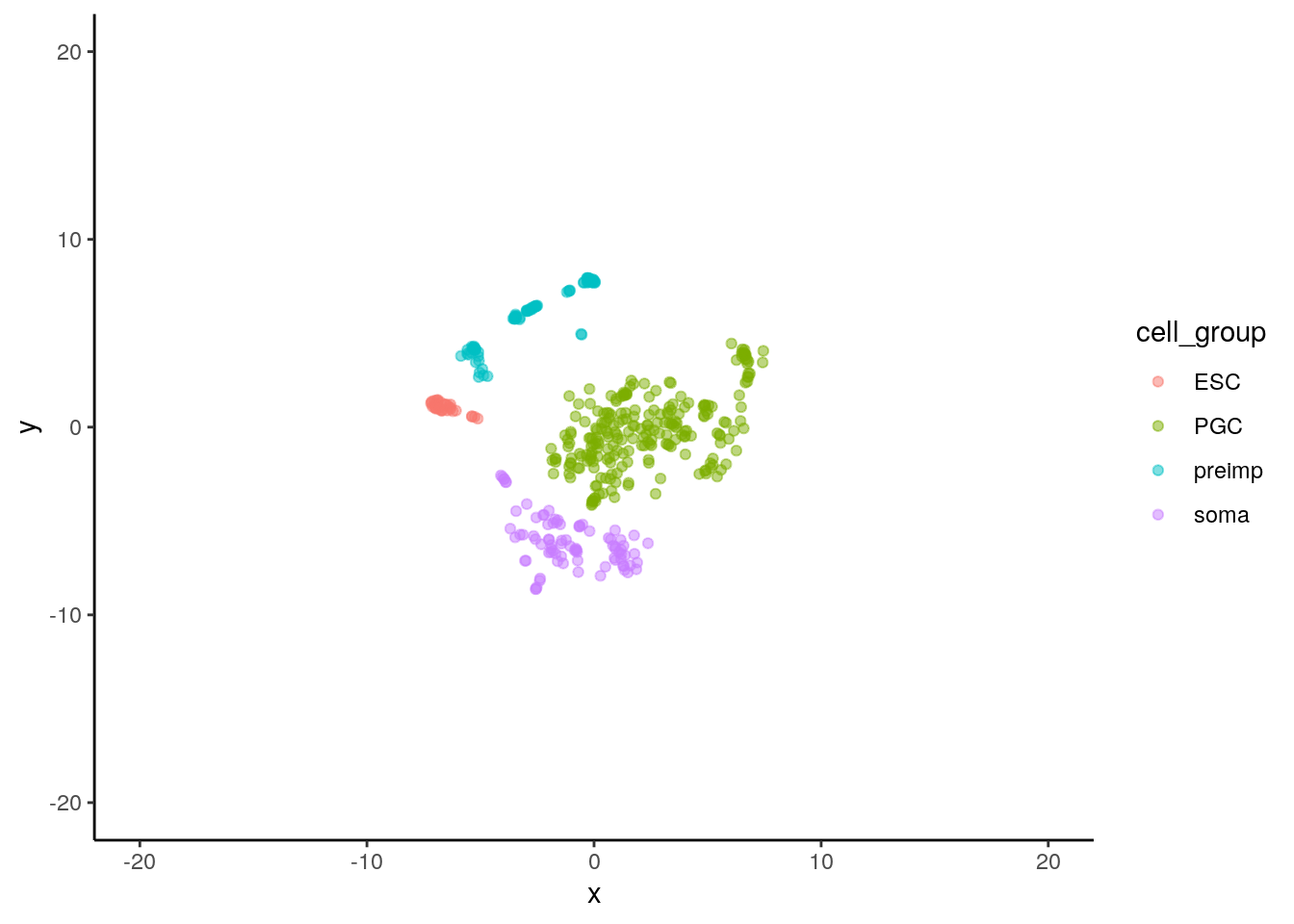
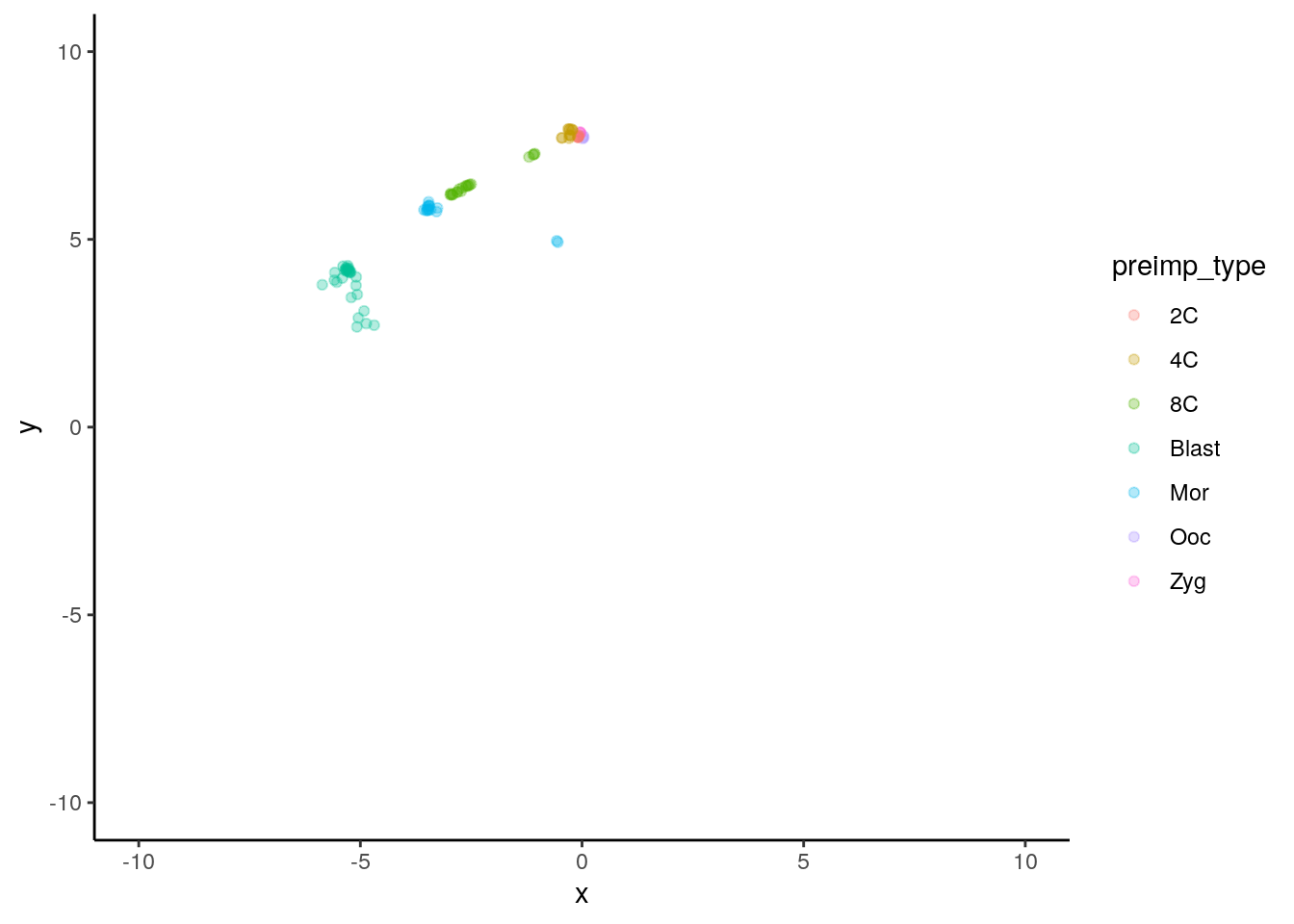
B.2 Exercise 2.
We can plot the expression patterns for pre-implantation embryos:
sc_2d_preimp <- inner_join(sc_2d_data, metadata, by='cell_type') %>%
filter(cell_group == 'preimp') %>%
mutate( preimp_type = recode(as.character(Time),
'0' = 'Ooc',
'1' = 'Zyg',
'2' = '2C',
'3' = '4C',
'4' = '8C',
'5' = 'Mor',
'6' = 'Blast'))
ggplot(data=sc_2d_preimp) +
geom_point( mapping = aes(x=x, y=y, color=preimp_type), alpha=0.3) +
scale_x_continuous(limits = c(-10,10)) +
scale_y_continuous(limits = c(-10,10)) +
theme_classic()