Homework before session 4
For our homework exercises, we will use a new dataset from the Gapminder Foundation, which gives access to global data as well as many tools to help explore it.
We will use data relating to socio-economic statistics for 2010. The columns in our data file are:
| Column | Description |
|---|---|
| country | country name |
| world_region | 6 world regions |
| year | year that each datapoint refers to |
| children_per_woman | total fertility rate |
| life_expectancy | average number of years a newborn child would live if current mortality patterns were to stay the same |
| income_per_person | gross domestic product per person adjusted for differences in purchasing power |
| is_oecd | Whether a country belongs to the “OECD” (TRUE) or not
(FALSE) |
| income_groups | categorical classification of income groups |
| population | total number of a country’s population |
| main_religion | religion of the majority of population in 2008 |
| child_mortality | death of children under 5 years old per 1000 births |
| life_expectancy_female | life expectancy at birth, females |
| life_expectancy_male | life expectancy at birth, males |
Task 1 - setting up your project
Tip: this exercise builds on the skills gained in Getting Started.
- On your computer, create a new directory for this project called
gapminder-dataviz. - Create directories that you feel are important, including one called
data_rawfor saving the raw data. - Download the gapminder socio-economic dataset 2010 and save it in your project’s data folder. (right-click the file link and choose “Save link as…”)
- Create a new R Project on the
gapminder-datavizdirectory you just created.
Answer
We create a new directory as well as sub-directories, shown here schematically:
gapminder-dataviz
|_ data_raw
|_ data_processed
|_ fig_output
|_ scriptsWe use data_raw to save the data file that we download
with the link provided.
Finally, we create an R project on this directory:
- Start RStudio.
- Under the File menu, click on New Project. Choose Existing Directory.
- Hit the Browse… button to go to the folder named
gapminder-datavizthat we’ve just created. - Click on Create Project.
RStudio should refresh itself and then indicate that the working
directory has been set to the new folder. For example, you can run
the command getwd() on the console to confirm that this is
the case.
Task 2 - importing data
As with any dataset you must first understand its content and formatting. Understanding what data you have will help you decide what story you can learn from the data and how best to present it
Create a new script to analyse these data and call it
01-gapminder_exploration.R. Then, populate it with code to
achieve the following:
- Read the
gapminder2010_socioeconomic.csvfile into adata.frame/tibbleobject calledgapminder.Hint
Use theread_csv()function. Remember to first load thetidyversepackage withlibrary(tidyverse). - Make a basic check that your data import went well. Your data frame
should have 193 rows and 13 columns.
Hint
Use functions such asnrow(),ncol(),summary()andstr()to check data integrity. - What types of variables does this data have?
- Are there any issues that you notice with these data? Note these down.
Answer
We can read our data as follows:
#> Rows: 193 Columns: 13
#> ── Column specification ──────────────────
#> Delimiter: ","
#> chr (5): country, world_region, income_groups, main_religion, life_expectanc...
#> dbl (7): year, children_per_woman, life_expectancy, income_per_person, popul...
#> lgl (1): is_oecd
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.To examine the contents of the data.frame we can use several functions, for example, to get the number of rows and columns:
ncol(gapminder) # number of columns in the data.frame
nrow(gapminder) # number of rows in the data.frameThe str() function gives a more comprehensive view of
the contents of the data.frame, including the number of rows and columns
as well as the type of variable each columns was imported as by R:
The summary function is also very useful as it gives a quick overview of the types of variables as well as average and quantiles for numeric data:
The types of variables we have are:
- nominal (categorical variable with no order): for example
countryandworld_region - ordinal (categorical variable with order): for example
income_groups - binary (categorical variable with two mutually exclusive values):
is_oecd - continuous (numerical variable with any values): for example
income_per_personorlife_expectancy - discrete (numerical variable with limited values only): for example
year
From the output of the summary function, we can notice a few issues with these data:
- Some
life_expectancy_malevalues are invalid == -999. life_expectancy_femalewas imported as a character variable, but should be numeric.- Some missing data - this is almost always expected in large datasets.
If we look at the top few rows of the table:
We can see that life_expectancy_female is showing the
value “-” in the 4th row of data. Probably the person recording these
data encoded missing values with the “-” symbol, but the
read_csv() function did not recognise this as missing data.
The default is to consider empty cells as missing data and so
we should correct this in the dataset to make sure all missing
values are encoded in the same way.
There are a few other issues in the main_religion
column, which were a little harder to detect. If we look at the unique
values of this column, we will notice different spellings/formats for
some of its values:
These types of spelling mistakes are very common and it’s important to be aware that R would consider “muslim” and “Muslim” to be different words (due to the case-sensitivity).
Task 3 - plotting
- Create a scatter plot for life expectancy (x) against income per person (y)
- You will notice you get a warning message here - why do you think this is?
Answer
#> Warning: Removed 6 rows containing missing values
#> (`geom_point()`).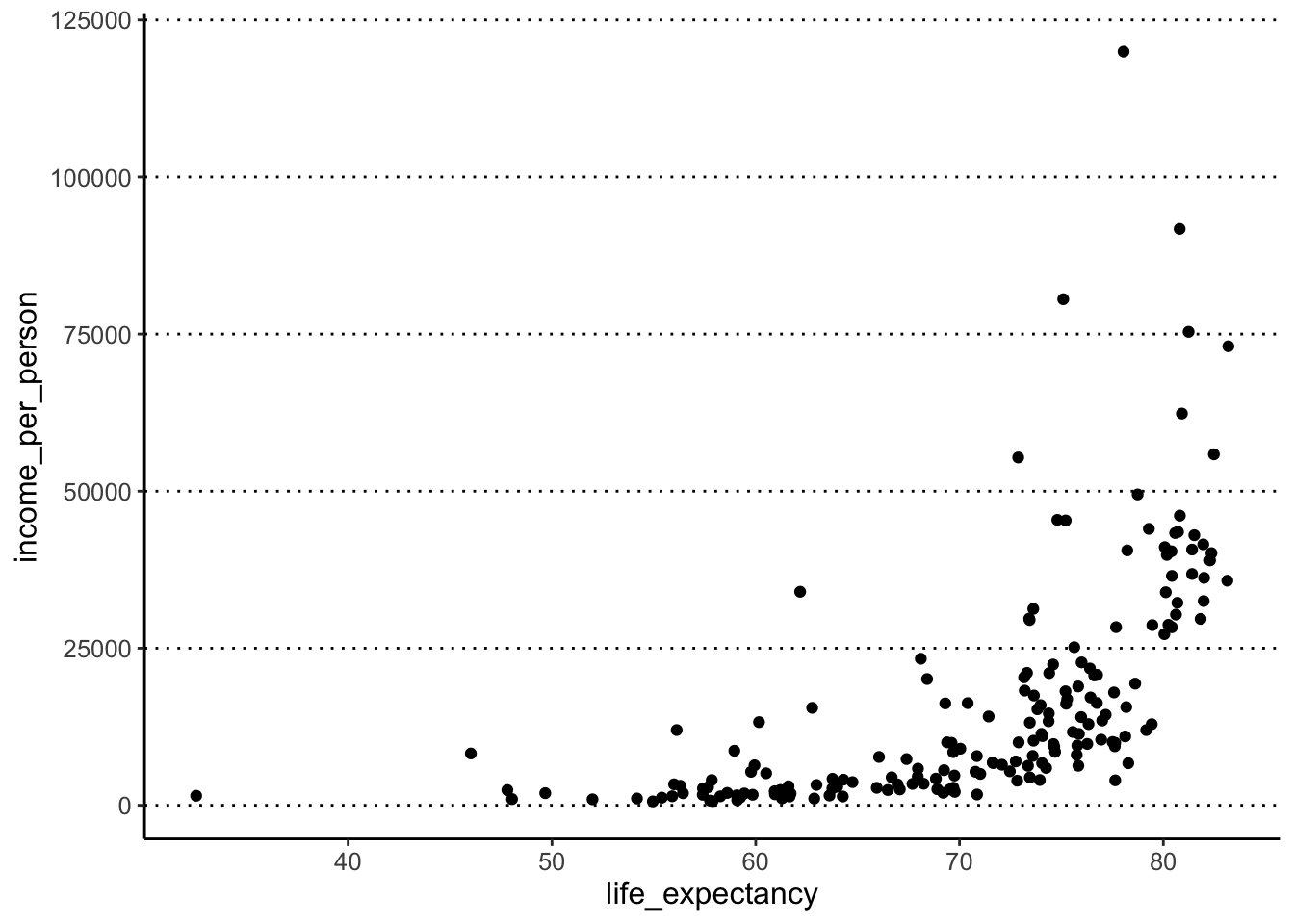
The warning message we get is because 6 of the rows in the data frame
do not have life_expectancy information (they are NA
missing data).
If it was a key variable for your analysis you might have wanted to remove those individuals with missing data. In this case, we don’t mind having these missing data, so we can carry on with our analysis.
Note that ‘Warning messages’ are simply that - a warning, not an error. They are very helpful and always worth reading.
Task 4 - data manipulation
- How many countries are there in “South Asia”?
Hint
Use thefilter()function to subset the table to retain only rows whereworld_region == "south_asia". - Create a new column in the table called
income_total, which is the product of population and income per person (i.e. the total average income of the country).Hint
Use themutate()function to create a new column.
Answer
To identify countries in South Asia, we can use the following:
From the output we can see this table has 8 rows, therefore 8 countries in this part of the world.
To create the new column we can use the mutate()
function, as such:
If we wanted to save this in our table, we need to update the object,
using the <- assignment:
