Starting with data
General concepts in data analysis
Before we start, I want to emphasise that even though the data used in this course are from an ecological study, the principals you learn can be applied to any analysis of tabular data.
Data exploration workflow
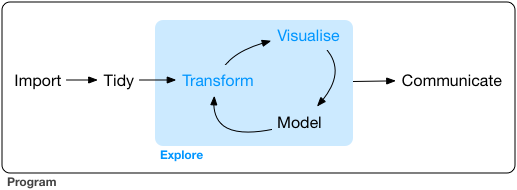
When you are working on a project that requires data analysis, you will normally need to perform the following steps:
 )
)
More information on this workflow can be found in the R for Data Science book. To understand better the workflow in the illustration above, let us go over each stage to see what each step entails:
- The first step in working with data is to first import your data into R. This connects the external file/database to your project in R.
- Cleaning or tidying the data will
follow, which involves making sure that the data is consistent and that
each row in the data set is an observation and each column is a
variable.
e.g. In the surveys data frame the month column specifies months as an integer from 1 to 12. The data set would have inconsistent data if there was a record in the data set that had a month specified by name, e.g. September rather than 9. A month of 0 or any other number that is not in the range 1 to 12 would have also made the data set inconsistent. Another common problem is capitalisation; the same word in the same column can be written with capitals or without; e.g. Bird or bird in the same taxa column is inconsistent data. During the tidying stage it is important to make the data set consistent and much as possible so that you can focus on the questions you are trying to solve in your analysis.
- Once the data set is tidy, we move to the transformation stage. To
be able to transform your data you need to plan in
advance what analyses you would like to perform on the data set and what
plots you would like to create. In this way, you are able to plan ahead
what variables/columns you will be using from the data set, what
additional variables you will need to create and what variables you will
not be using so that you can keep only the columns in the data set that
are relevant for your analyses. By the end of the transformation process
you will have a data set that is focused for your analyses and you can
move on to the main exploratory mechanisms of this workflow which are
visualisation and modelling. These two stages complement each other and
when exploring your data you normally repeat these two stages several
times.
- Visualising data is a powerful way to explore your
data. Furthermore it helps you understand if there is any pattern in the
data.
- Modelling the data involves applying statistics or
other mathematical or computational models on your data to explore if
there are correlations or patterns in the data set to help you answer
the scientific question you are trying to solve.
- The last step in the data exploration workflow is to communicate your results. This is very important as you will need to be able to communicate your results to others to have a successful project.
All these stages in the data exploration workflow can be achieved by programming in R. In these sessions we will not look into the Model and Communicate stages of the workflow, but there are specialised courses available here at the Bioinformatics Training Facility that cover those topics. If you want to know more, see:
- Model: Statistics for Biologists in R and An Introduction to Machine Learning
- Communicate: Reproducible Research with R
In the next sections we will be looking at the import, tidy, transform and visualise stages of the data exploration workflow by using one of the most popular packages in data science in R; tidyverse. We introduced this in the previous session and now we will see a lot more of its functionality.
Understanding data
To do in-depth data analyses, it is crucial you understand your data. So before we start doing any form of analyses we will first try to understand the data set that we will be using throughout this course. Let us first download the file and have a look at the data.
Thinking back to the structure of our R project, we have a working directory. Within the working directory we can create folders to organise our files. We are going to download some raw data and it is good practice to keep your raw data separate from other data, because that way you can always refer back to the data that you started with.
In this case we have already generated the relevant folders in the Getting started section, but in case you still need to do this, here is a reminder:
You can create folders straight from RStudio from the right bottom pane in the Files section > New Folder icon.
Remember to try and avoid capitalisation and spaces (use the underscore instead).
We are now ready to download the data, using the R function
download.file() to download the CSV file that contains the
data.
download.file(url="https://ndownloader.figshare.com/files/2292169",
destfile = "data_raw/portal_data_joined.csv")Inside the download.file command, the first entry is a character string with the source URL (“https://ndownloader.figshare.com/files/2292169”). This source URL downloads a CSV file from figshare. The text after the comma (“data_raw/portal_data_joined.csv”) is the destination of the file on your local machine.
The data set has the following columns, with each row holding information for a single animal:
| Column | Description |
|---|---|
| record_id | Unique id for the observation |
| month | month of observation |
| day | day of observation |
| year | year of observation |
| plot_id | ID of a particular plot |
| species_id | 2-letter code |
| sex | sex of animal (“M”, “F”) |
| hindfoot_length | length of the hindfoot in mm |
| weight | weight of the animal in grams |
| genus | genus of animal |
| species | species of animal |
| taxon | e.g. Rodent, Reptile, Bird, Rabbit |
| plot_type | type of plot |
Reading in data from a file
Next we need to load the data into R and look at how the
data is loaded into R. We will use read_csv() from the
tidyverse package to load into memory the content of the
CSV file.
We already installed and loaded the tidyverse library,
but if you have not, then use install.packages("tidyverse")
to install it and library(tidyverse) to load it.
You can load the data using the following command:
#> Rows: 34786 Columns: 13
#> ── Column specification ──────────────────
#> Delimiter: ","
#> chr (6): species_id, sex, genus, species, taxa, plot_type
#> dbl (7): record_id, month, day, year, plot_id, hindfoot_length, weight
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.What this does is to use the read_csv() function to read
in the CSV file that contains our data. the data should be in a
data_raw folder, within your working directory. It then
assigns the data to an object called surveys.
The statement doesn’t produce any output because, as you might
recall, because assignments don’t display anything. It does give you
information on how the data was loaded. Note that some columns are
classed as col_double (numbers) and others as
col_character (text). This can be useful and important
information, because it tells you the type of data R considers it to be.
It also helps you check your data. For example, if R would view a column
as col_character but you know that it should only contain
numbers, then you know that some of the data are probably text, meaning
there are errors in your data that need a closer look.
If we want to find out how our data has been loaded, we can visualise
the contents of the data frame by typing its name
surveys:
#> # A tibble: 34,786 × 13
#> record_id month day year plot_id species_id sex hindfoot_length weight
#> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <dbl> <dbl>
#> 1 1 7 16 1977 2 NL M 32 NA
#> 2 72 8 19 1977 2 NL M 31 NA
#> 3 224 9 13 1977 2 NL <NA> NA NA
#> 4 266 10 16 1977 2 NL <NA> NA NA
#> 5 349 11 12 1977 2 NL <NA> NA NA
#> 6 363 11 12 1977 2 NL <NA> NA NA
#> 7 435 12 10 1977 2 NL <NA> NA NA
#> 8 506 1 8 1978 2 NL <NA> NA NA
#> 9 588 2 18 1978 2 NL M NA 218
#> 10 661 3 11 1978 2 NL <NA> NA NA
#> # ℹ 34,776 more rows
#> # ℹ 4 more variables: genus <chr>, species <chr>, taxa <chr>, plot_type <chr>The first line of the output shows the data
structure used to store the data imported into:
tibble. A tibble is the main
data structure used in tidyverse. You can look at
tibble as the data.frame version of
tidyverse. The first immediate difference from a
data.frame is that a tibble displays the data
type of each column under its name (handy) and it only prints as many
columns as fit on one screen (even handier, otherwise it would print
34,786 rows!).
Exercise - Understanding data
Try to do the following:
- Find out what data types there are in
surveys(numeric, categorical). - Are there missing values in the data? If so, can you see how many?
Hint: you can use the summary()
function to get more information on your data set
Answer
Using the summary() function gives us information about
each variable with summary metrics (such as the average and quartiles of
numeric variables). It also gives us information about missing values
(NA’s).
For example, from this output we can see that we have 2503 missing values for “weight” and 3348 missing values for “hindfoot_length”.
Data frames
Data frames are one of the most widely used type of data structure in R. It is very popular as most of the data is readily available in tabular form and it is the also the data structure used when plotting and performing most analyses in R.
A data frame can be compared to what you would see in an Excel spreadsheet: a rectangular data set.
A data frame is the representation of data in the format of a table where the columns are vectors that all have the same length. Because columns are vectors, each column must contain a single type of data (e.g., characters, integers, logical). For example, here is a figure depicting a data frame comprising a numeric, a character, and a logical vector.
We can access the values of a single column of the data.frame using
the $ notation, as such:
The result is a vector (not a data.frame), similar to what
we created using the c() function in the Introduction to R section.
Therefore we can use functions such as the ones we used with vectors, for example to calculate the mean of this variable:
Now that we have loaded our data into R and understand its underlying structure, we can move on to doing some analysis!
Creating plots
One of the best ways of looking at your data is to visualise them.
The function that helps you visualise your data is called
ggplot().
The basic format for using ggplot() is as follows:
ggplot(data = <DATA>, mapping = aes(<MAPPINGS>)) + <GEOM_FUNCTION>()As you can see, there are 3 main elements that you need to create a plot:
The ggplot function takes 2 arguments:
- data: This is the data frame to attach to the plot. The data frame must contain the variables to plot as columns and the rows must contain the observations that you need to plot.
- mapping: Aesthetic mappings describe how variables in the data are mapped to visual properties of the plot.
Using the ggplot function on its own will not plot
anything. We need to add a geom_function as a layer.
Layers are added to plots by using +. They are added on top
of the other previous layers that might be present.
- geom_function: This specifies the type of plot would you like to plot. The greatest advantage of this is that you can easily change the plot type by just changing the geom_function and keeping everything else the same. You can see a whole list of plots that you can plot here.
Let’s practice this on our surveys data set. We would
like to create a scatter plot with weight on the x-axis,
hindfoot_length on the y-axis:
Adding layers
If you just specify the ggplot function with the data
and aesthetic mappings, it will create a plot, but the data itself is
not displayed. Let us now add the geom_function for the
scatter plot (geom_point()) as a layer to the plot:
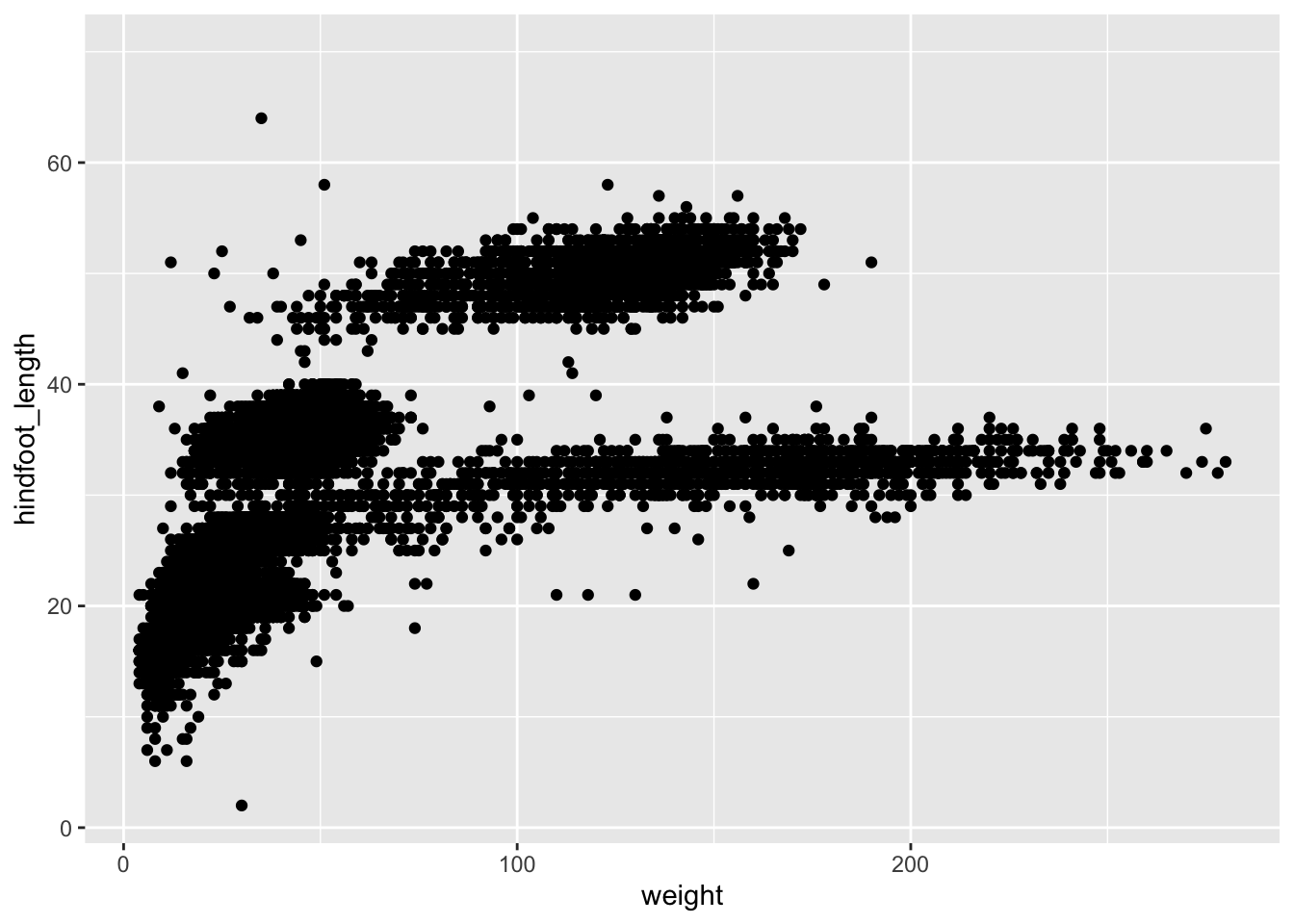
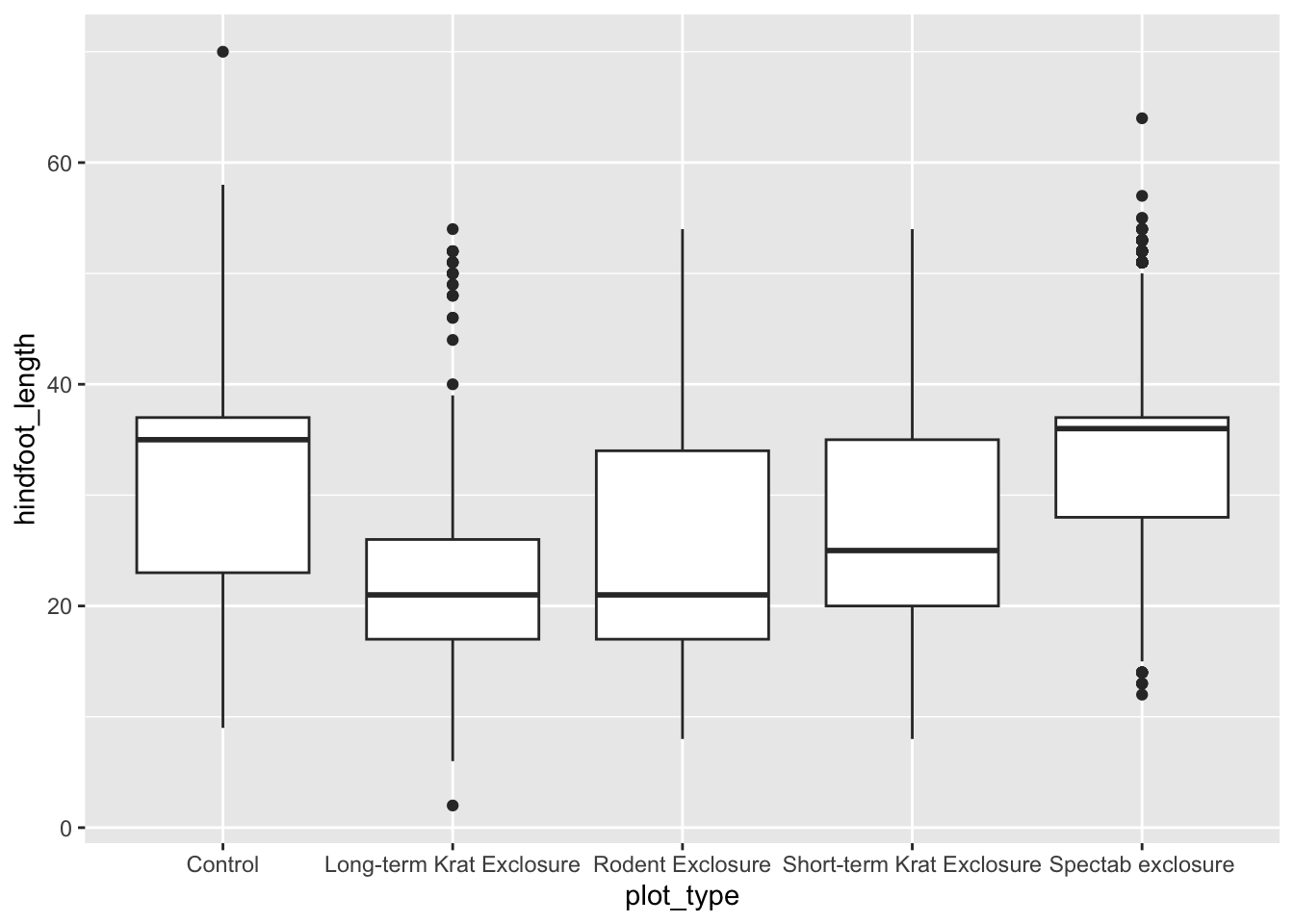
Exercise - Boxplot
The plot we created above was a simple scatter plot, which used the
geom_point() function. There are many different types of
geoms in ggplot and to explore this we would like you to create a box plot,
using the same data set.
- The geoms are named quite logically, so finding the right geom for a box plot should be OK
- The scatter plot uses two continuous variables. A box plot should
only have a continuous y-variable, so on the x-axis you should plot a
categorical variable (think back of your
summary()output!)
Transforming data - dplyr
In most of the cases you will need to change the format of your
dataset because it will not be in the right format that you will need to
plot or analyse the data. tidyverse has a package called
dplyr which contains functions that help you to select
columns/rows, sort, combine and perform other data types of data
transformations. In the next sections we will look at different ways to
transform our dataset. Now that we already know the basics of
visualising data with ggplot we will also learn how to
visualise other plots with the transformed dataset as we go along.
To learn more about dplyr please look at the following
resources:
If you find these resources difficult to understand, return to these after completing the course.
Pipes
Before we move onto the different ways that we can transform our data, we’ll discuss one of the most powerful additions to R, pipes.
Pipes let you take the output of one function and send it directly to
the next, which is useful when you need to do many things to the same
dataset. Pipes in R look like %>% and are made available
via the magrittr package, installed
automatically with dplyr.
Let’s say we are interested only in the data that do not contain any missing values. Using pipes, we do the following:
This takes the argument on the left (surveys) and passes
it on to the function after it (drop_na, which removes all
rows with missing values).
If we want to create a new object with the transformed data we can assign it a new name as below:
Although at this point pipes might not seem like a revolutionary invention, they become more powerful when combining multiple operations. This we’ll see next.
Selecting columns
To select columns of a data frame or tibble, use
theselect function. The first argument is the data frame or
tibble you are working on (in our example it is surveys,
which we pipe through), and the subsequent arguments are the columns to
keep.
# Extract species_id, weight, hindfoot_lenth, year and sex columns from surveys dataset.
surveys %>%
select(species_id, weight, hindfoot_length, year, sex)To select all columns except certain ones, put a
- in front of the column to exclude it.
# Select all columns of the surveys dataset apart from record_id and species_id columns.
surveys %>%
select(-record_id, -species_id)Filtering rows
To remove rows from a data frame or tibble use the
filter function from the dplyr package. The
first argument is the data frame or tibble to perform the filtering on
(in this case we pipe the data through) and the next arguments are the
conditions on which to keep the rows.
Filtering rows by values
To choose rows based on a specific condition, use the
filter function as follows:
# Keep only the observations of animals collected from 1995 onwards from the surveys dataset.
surveys %>%
filter(year >= 1995)You can filter on multiple conditions:
# Keep only the observations of animals collected from 1995 onwards
# that are female from the surveys dataset.
surveys %>%
filter(year >=1995,
sex == "F")To quote or not to quote?
The sex column is a character and thus needs to be
quoted, whereas the year column is numerical and does not.
Also note that the filter arguments could have been written on a single
line, but it is useful to break up your code sometimes to make it more
readable.
Exercise - Pipes
Subset the surveys_complete data to keep only the
species_id, weight,
hindfoot_length, year and sex
columns and the animals collected on and after 1995. How many rows are
left?
Creating new columns
Often you’ll want to create new columns based on the values in
existing columns, for example to do unit conversions, or to find the
ratio of values in two columns. For this we’ll use the
mutate function.
To create a new column of weight in kg:
You can also create a second new column based on the first new column
within the same call of mutate():
There are other ways on how to create new columns. Refer to the dplyr cheat sheet Make New Variables section.