# Load required R libraries
library(tidyverse)
library(broom)
# Read in the required data
challenger_data <- read_csv("data/challenger.csv")8 Proportional response
- Be able to analyse proportional response variables
- Be able to create a logistic model to test proportional response variables
- Be able to plot the data and fitted curve
8.1 Context
In the previous section, we explored how logistic regression can be used to model binary outcomes. We can extend this approach to proportional outcomes — situations where we count how many times an event occurs out of a fixed number of trials (for example, the number of successes out of 10 attempts).
In this section, we illustrate how to model proportional data using logistic regression, using the well-known Challenger dataset as an example.
8.2 Section setup
We’ll use the following libraries and data:
# Load required Python libraries
import pandas as pd
import math
import statsmodels.api as sm
import statsmodels.formula.api as smf
from scipy.stats import *
from plotnine import *
# Read in the required data
challenger_data = pd.read_csv("data/challenger.csv")8.3 The Challenger dataset
The example in this section uses the challenger data. These data, obtained from the faraway package in R, contain information related to the explosion of the space shuttle Challenger on 28 January, 1986. An investigation traced the probable cause back to certain joints on one of the two solid booster rockets, each containing six O-rings that ensured no exhaust gases could escape from the booster.
The night before the launch was unusually cold, with temperatures below freezing. The final report suggested that the cold snap during the night made the o-rings stiff and unable to adjust to changes in pressure - leading to a proportion of them failing during launch. As a result, exhaust gases leaked away from the solid booster rockets, causing one of them to break loose and rupture the main fuel tank, leading to the final explosion.
The question we’re trying to answer in this session is: based on the data from the previous flights, would it have been possible to predict the failure of most o-rings on the Challenger flight?
8.4 Load and visualise the data
First we load the data, then we visualise it.
challenger <- read_csv("data/challenger.csv")Rows: 23 Columns: 2
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
dbl (2): temp, damage
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.challenger_py = pd.read_csv("data/challenger.csv")The data set contains several columns:
temp, the launch temperature in degrees Fahrenheitdamage, the number of o-rings that showed erosion
Before we have a further look at the data, let’s calculate the proportion of damaged o-rings (prop_damaged) and the total number of o-rings (total) and update our data set.
challenger <-
challenger |>
mutate(total = 6, # total number of o-rings
intact = 6 - damage, # number of undamaged o-rings
prop_damaged = damage / total) # proportion damaged o-rings
challenger# A tibble: 23 × 5
temp damage total intact prop_damaged
<dbl> <dbl> <dbl> <dbl> <dbl>
1 53 5 6 1 0.833
2 57 1 6 5 0.167
3 58 1 6 5 0.167
4 63 1 6 5 0.167
5 66 0 6 6 0
6 67 0 6 6 0
7 67 0 6 6 0
8 67 0 6 6 0
9 68 0 6 6 0
10 69 0 6 6 0
# ℹ 13 more rowschallenger_py['total'] = 6
challenger_py['intact'] = challenger_py['total'] - challenger_py['damage']
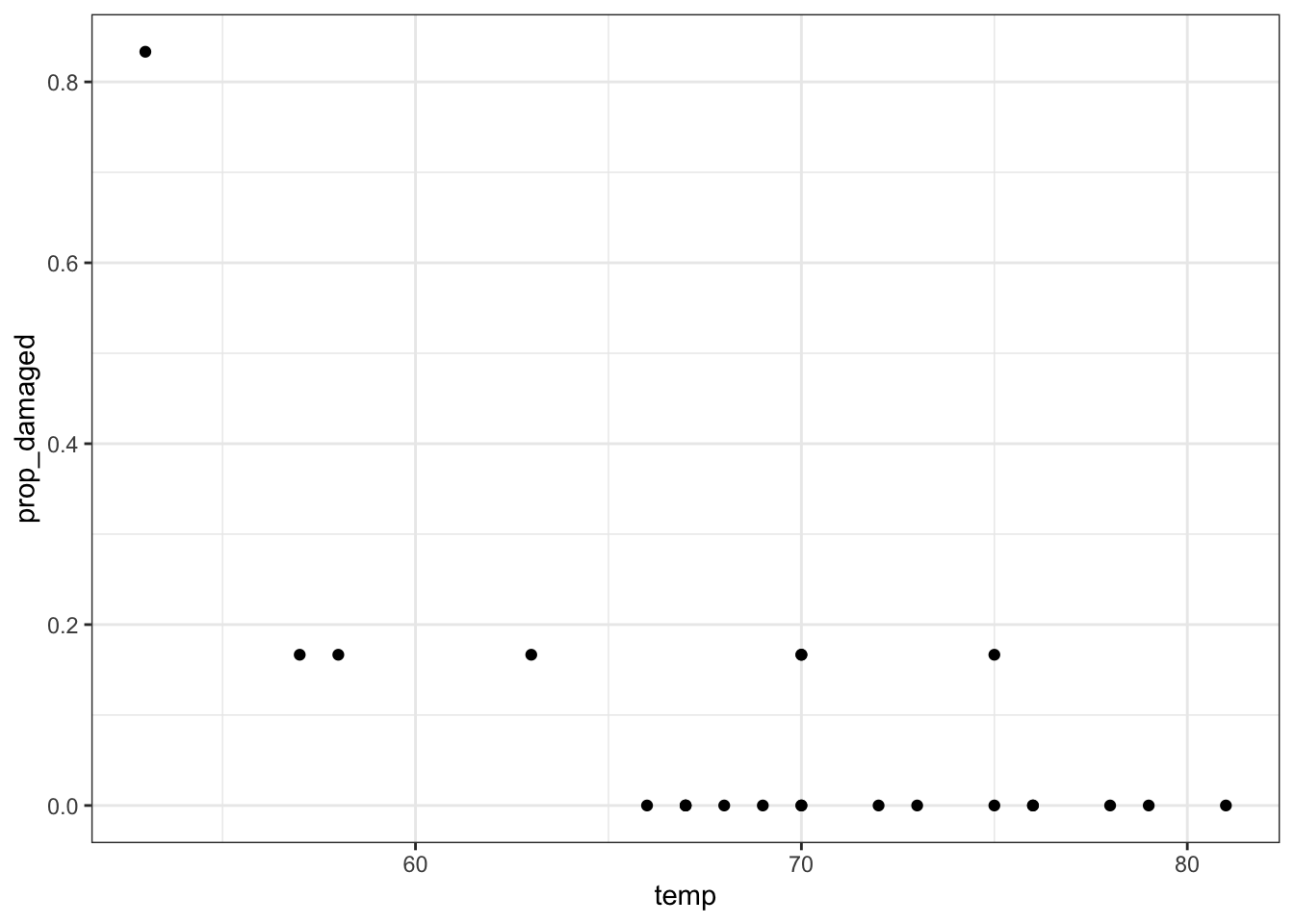
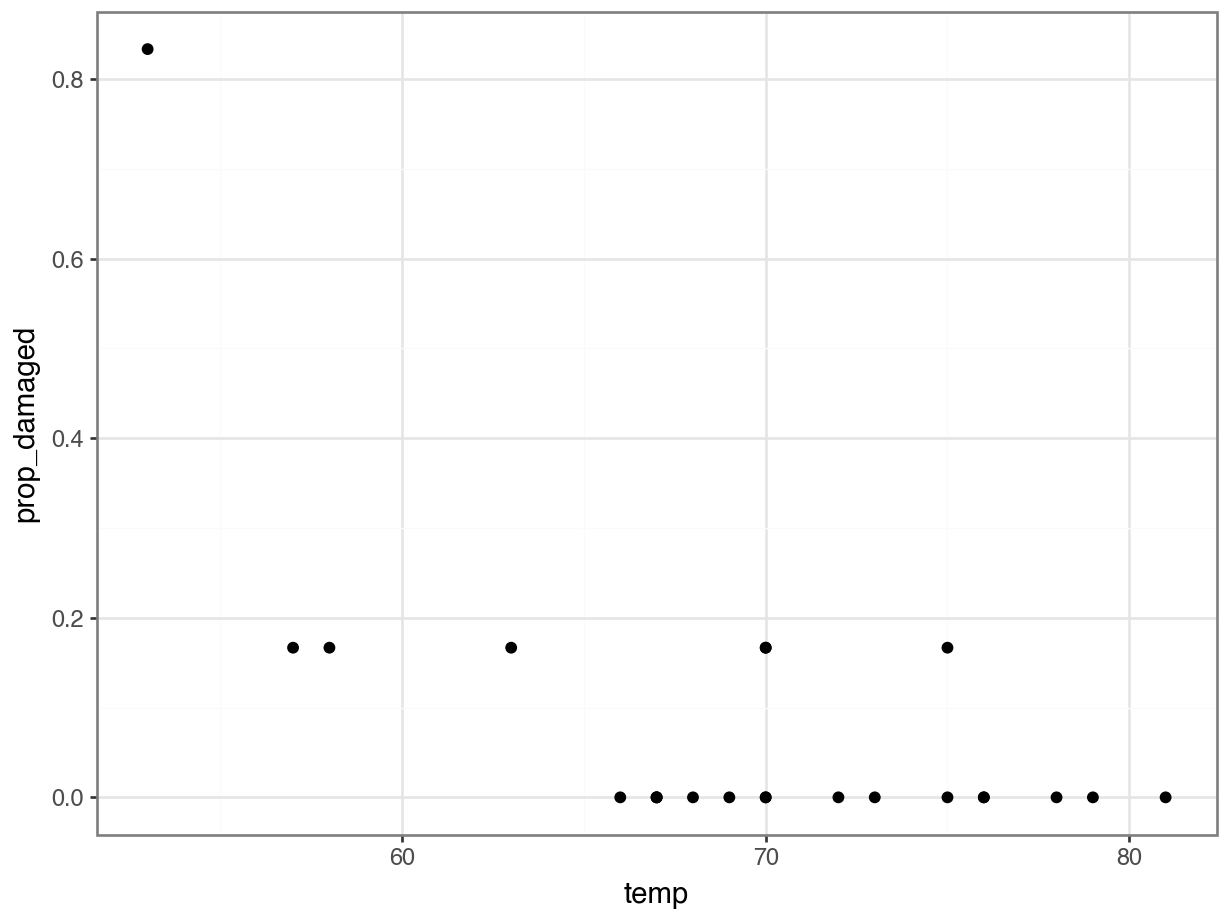
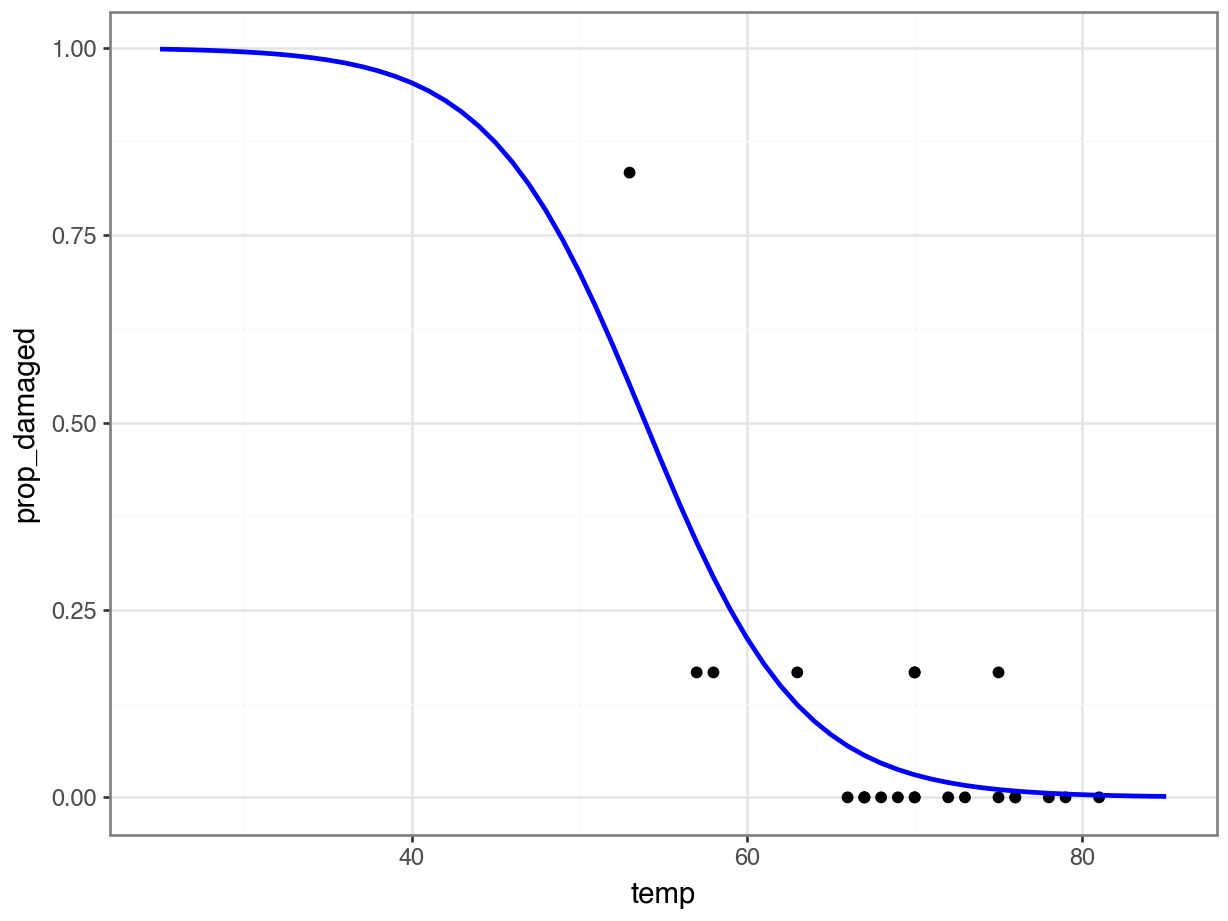
challenger_py['prop_damaged'] = challenger_py['damage'] / challenger_py['total']Plotting the proportion of damaged o-rings against the launch temperature shows the following picture:
The point on the left is the data point corresponding to the coldest flight experienced before the disaster, where five damaged o-rings were found. Fortunately, this did not result in a disaster.
Here we’ll explore if we could have reasonably predicted the failure of both o-rings on the Challenger flight, where the launch temperature was 31 degrees Fahrenheit.
8.5 Creating a suitable model
We only have 23 data points in total. So we’re building a model on not that much data - we should keep this in mind when we draw our conclusions!
We are using a logistic regression for a proportion response in this case, since we’re interested in the proportion of o-rings that are damaged.
We can define this as follows:
glm_chl <- glm(cbind(damage, intact) ~ temp,
family = binomial,
data = challenger)Defining the relationship for proportion responses is a bit annoying, where you have to give the glm model a two-column matrix to specify the response variable.
Here, the first column corresponds to the number of damaged o-rings, whereas the second column refers to the number of intact o-rings. We use the cbind() function to bind these two together into a matrix.
# create a generalised linear model
model = smf.glm(formula = "damage + intact ~ temp",
family = sm.families.Binomial(),
data = challenger_py)
# and get the fitted parameters of the model
glm_chl_py = model.fit()If we’re using the original observations, we need to supply both the number of damaged o-rings and the number of intact ones.
prop_damaged as our response variable?
You might have noticed that when we write our model formula, we are specifically writing cbind(damage, intact) (R) or damage + intact (Python), instead of the prop_damaged variable we used for plotting.
This is very deliberate - it’s absolutely essential we do this, in order to fit the correct model.
When modelling our data, we need to provide the function with both the number of successes (damaged o-rings) and the number of failures (intact o-rings) - and therefore, implicitly, the total number of o-rings - in order to properly model our proportional variable.
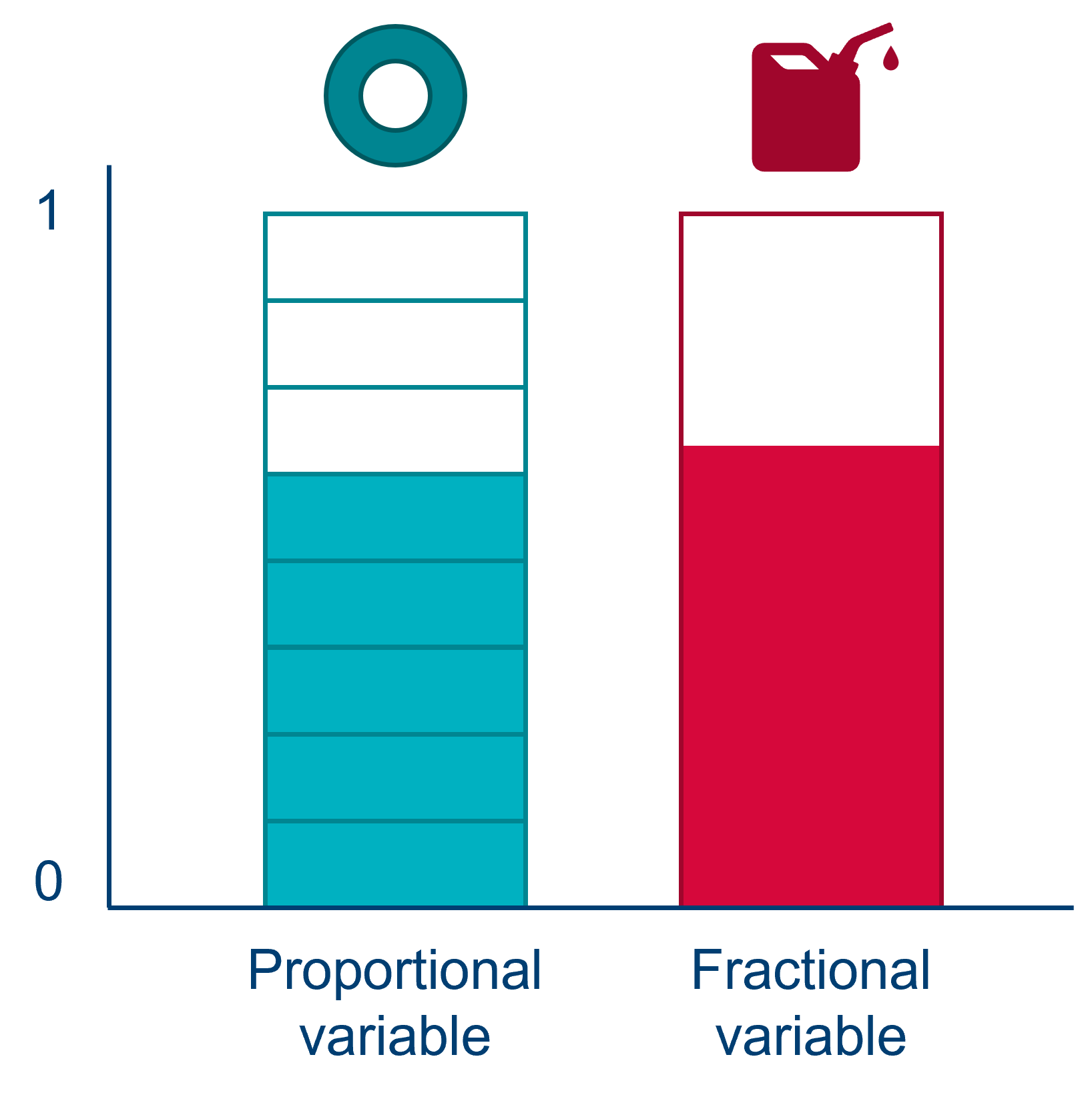
If we provide it with just the prop_damaged, then R/Python will incorrectly think our response variable is fractional.
A fractional variable is not constructed from a series of trials with successes/failures. It doesn’t have trials - there’s just a single value somewhere between 0 and 1 (or a percentage). An example of a fractional response variable might be what percentage of rocket fuel was used on a launch. We would not model this response variable with a logistic regression.
8.6 Model output
That’s the easy part done! The trickier part is interpreting the output. First of all, we’ll get some summary information.
Next, we can have a closer look at the results:
summary(glm_chl)
Call:
glm(formula = cbind(damage, intact) ~ temp, family = binomial,
data = challenger)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 11.66299 3.29626 3.538 0.000403 ***
temp -0.21623 0.05318 -4.066 4.78e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 38.898 on 22 degrees of freedom
Residual deviance: 16.912 on 21 degrees of freedom
AIC: 33.675
Number of Fisher Scoring iterations: 6We can see that the p-values of the intercept and temp are significant. We can also use the intercept and temp coefficients to construct the logistic equation, which we can use to sketch the logistic curve.
print(glm_chl_py.summary()) Generalized Linear Model Regression Results
================================================================================
Dep. Variable: ['damage', 'intact'] No. Observations: 23
Model: GLM Df Residuals: 21
Model Family: Binomial Df Model: 1
Link Function: Logit Scale: 1.0000
Method: IRLS Log-Likelihood: -14.837
Date: Fri, 25 Jul 2025 Deviance: 16.912
Time: 08:49:44 Pearson chi2: 28.1
No. Iterations: 7 Pseudo R-squ. (CS): 0.6155
Covariance Type: nonrobust
==============================================================================
coef std err z P>|z| [0.025 0.975]
------------------------------------------------------------------------------
Intercept 11.6630 3.296 3.538 0.000 5.202 18.124
temp -0.2162 0.053 -4.066 0.000 -0.320 -0.112
==============================================================================\[E(prop \ failed\ orings) = \frac{\exp{(11.66 - 0.22 \times temp)}}{1 + \exp{(11.66 - 0.22 \times temp)}}\]
Let’s see how well our model would have performed if we would have fed it the data from the ill-fated Challenger launch.
ggplot(challenger, aes(temp, prop_damaged)) +
geom_point() +
geom_smooth(method = "glm", se = FALSE, fullrange = TRUE,
method.args = list(family = binomial)) +
xlim(25,85)Warning in eval(family$initialize): non-integer #successes in a binomial glm!We get the warning Warning in eval(family$initialize): non-integer #successes in a binomial glm!. This is because we’re feeding a proportion value between 0 and 1 to a binomial GLM. What it is expecting is the number of successes out of a fixed number of trials. See the explanation above for more information. We’ll revisit this later!
We can get the predicted values for the model as follows:
challenger_py['predicted_values'] = glm_chl_py.predict()
challenger_py.head() temp damage total intact prop_damaged predicted_values
0 53 5 6 1 0.833333 0.550479
1 57 1 6 5 0.166667 0.340217
2 58 1 6 5 0.166667 0.293476
3 63 1 6 5 0.166667 0.123496
4 66 0 6 6 0.000000 0.068598This would only give us the predicted values for the data we already have. Instead we want to extrapolate to what would have been predicted for a wider range of temperatures. Here, we use a range of \([25, 85]\) degrees Fahrenheit.
model = pd.DataFrame({'temp': list(range(25, 86))})
model["pred"] = glm_chl_py.predict(model)
model.head() temp pred
0 25 0.998087
1 26 0.997626
2 27 0.997055
3 28 0.996347
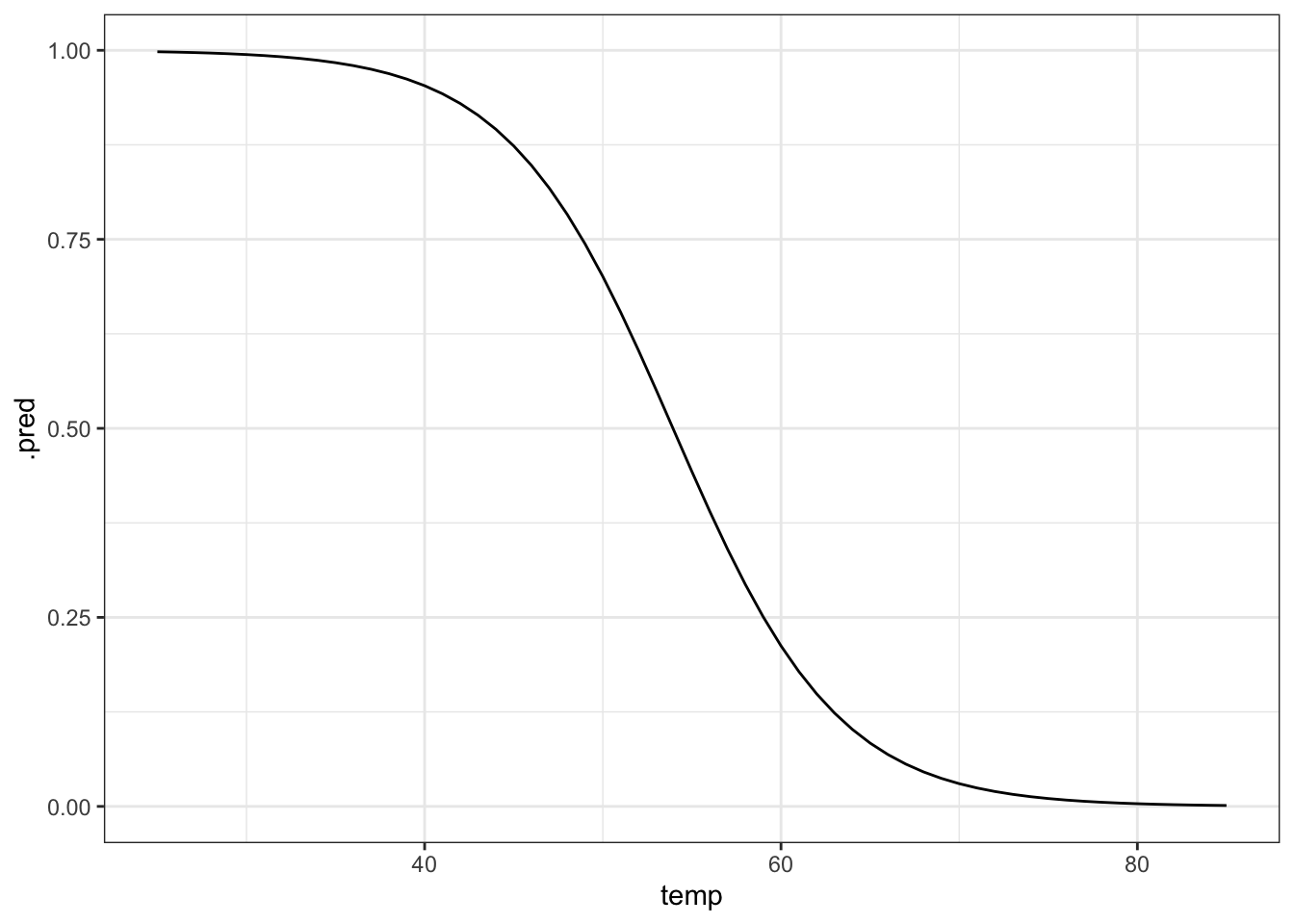
4 29 0.995469Another way of doing this it to generate a table with data for a range of temperatures, from 25 to 85 degrees Fahrenheit, in steps of 1. We can then use these data to generate the logistic curve, based on the fitted model.
# create a table with sequential numbers ranging from 25 to 85
new_data <- tibble(temp = seq(25, 85, by = 1))
model <- new_data |>
# add a new column containing the predicted values
mutate(.pred = predict(glm_chl, newdata = new_data, type = "response"))
ggplot(model, aes(temp, .pred)) +
geom_line()# plot the curve and the original data
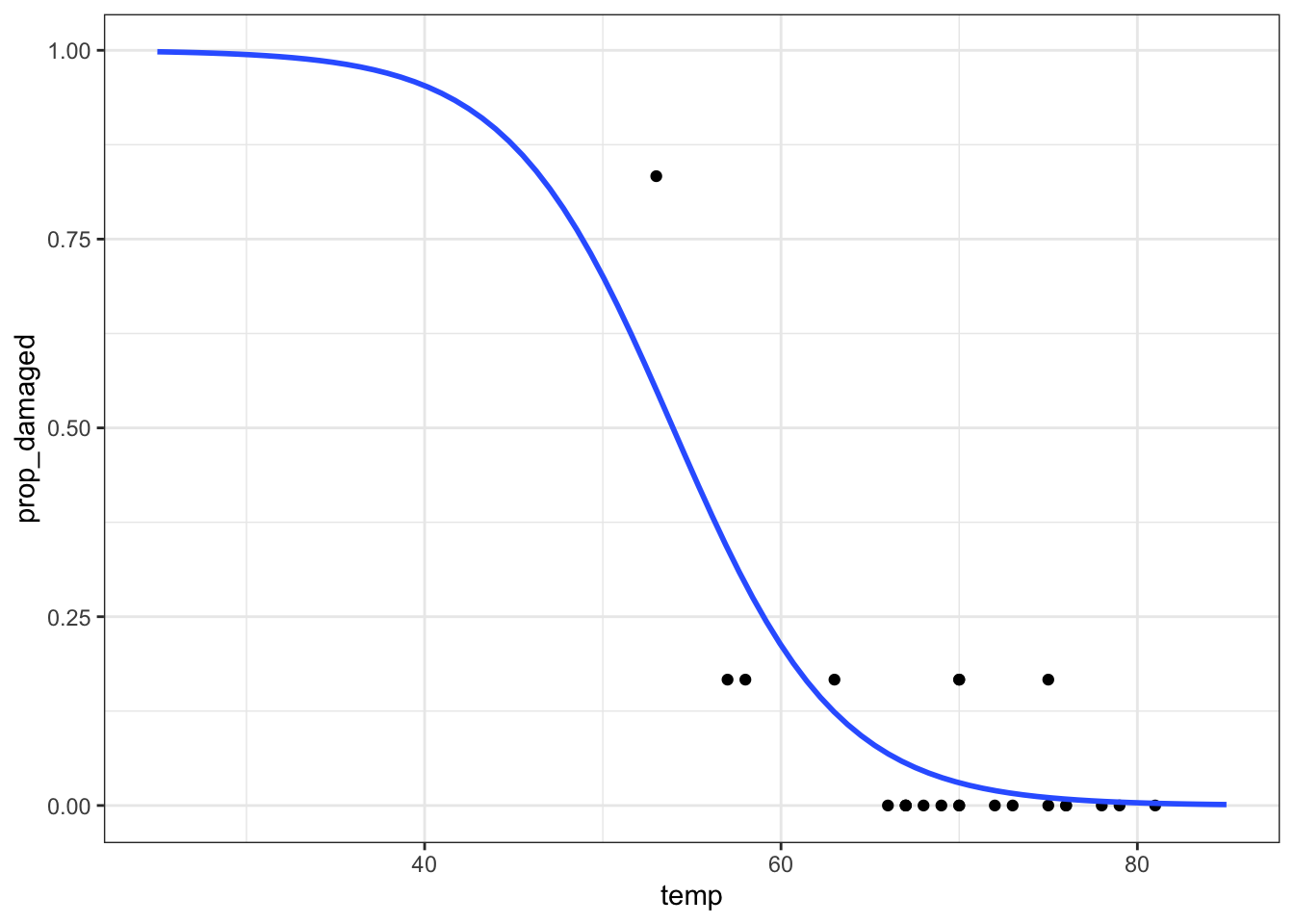
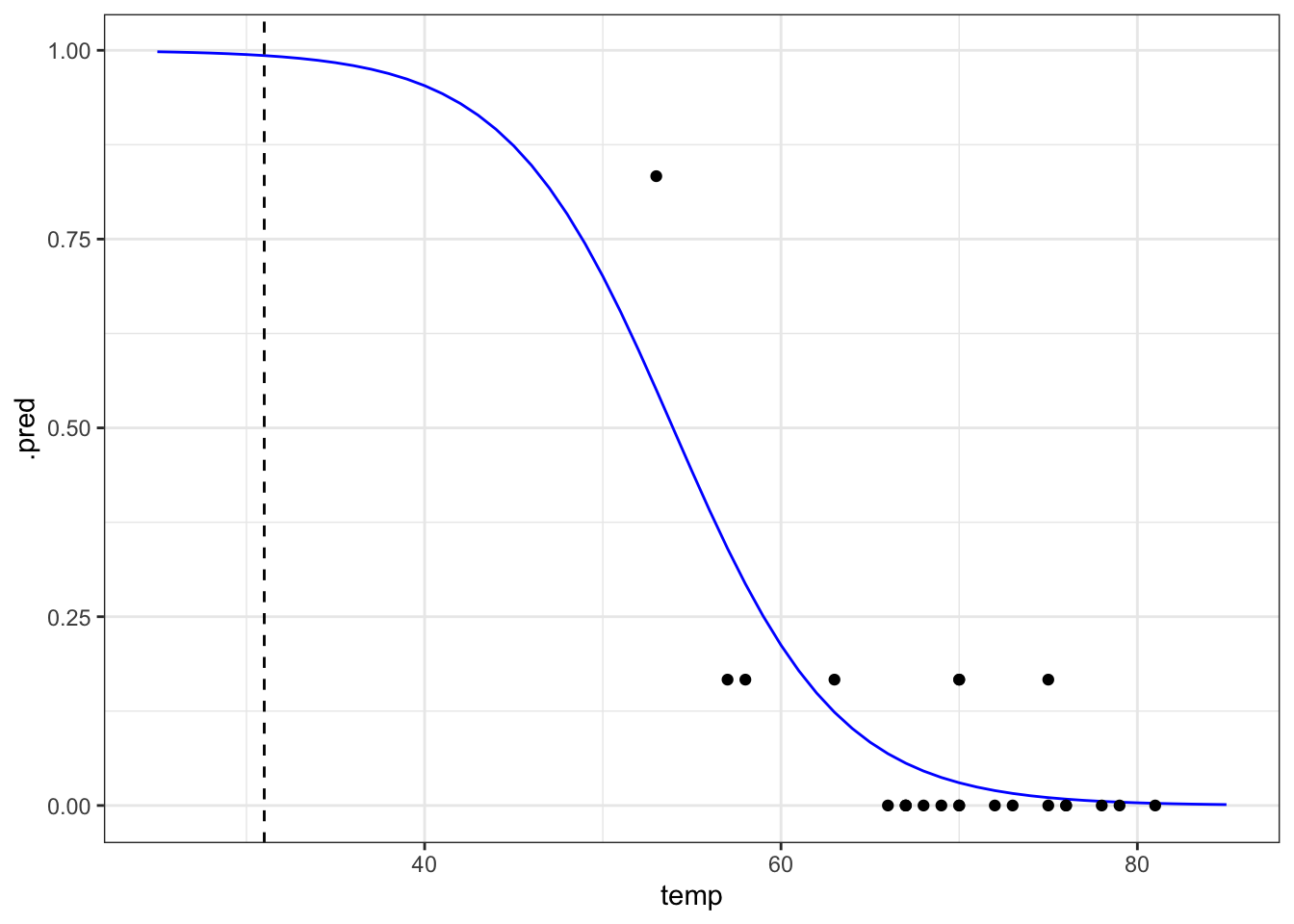
ggplot(model, aes(temp, .pred)) +
geom_line(colour = "blue") +
geom_point(data = challenger, aes(temp, prop_damaged)) +
# add a vertical line at the disaster launch temperature
geom_vline(xintercept = 31, linetype = "dashed")It seems that there was a high probability of both o-rings failing at that launch temperature. One thing that the graph shows is that there is a lot of uncertainty involved in this model. We can tell, because the fit of the line is very poor at the lower temperature range. There is just very little data to work on, with the data point at 53 F having a large influence on the fit.
We already did this above, since this is the most straightforward way of plotting the model in Python.
8.7 Exercises
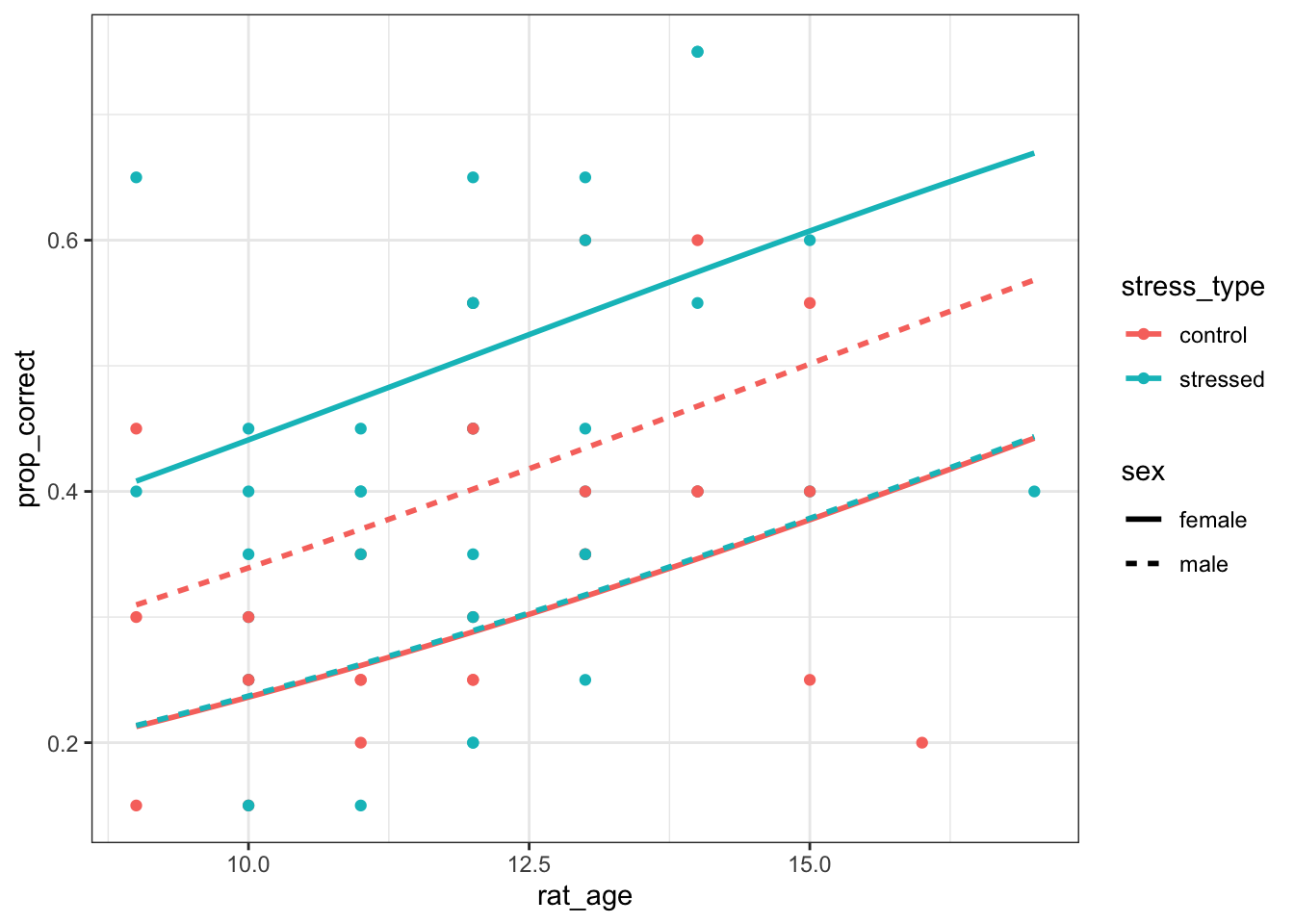
8.7.1 Rats and levers
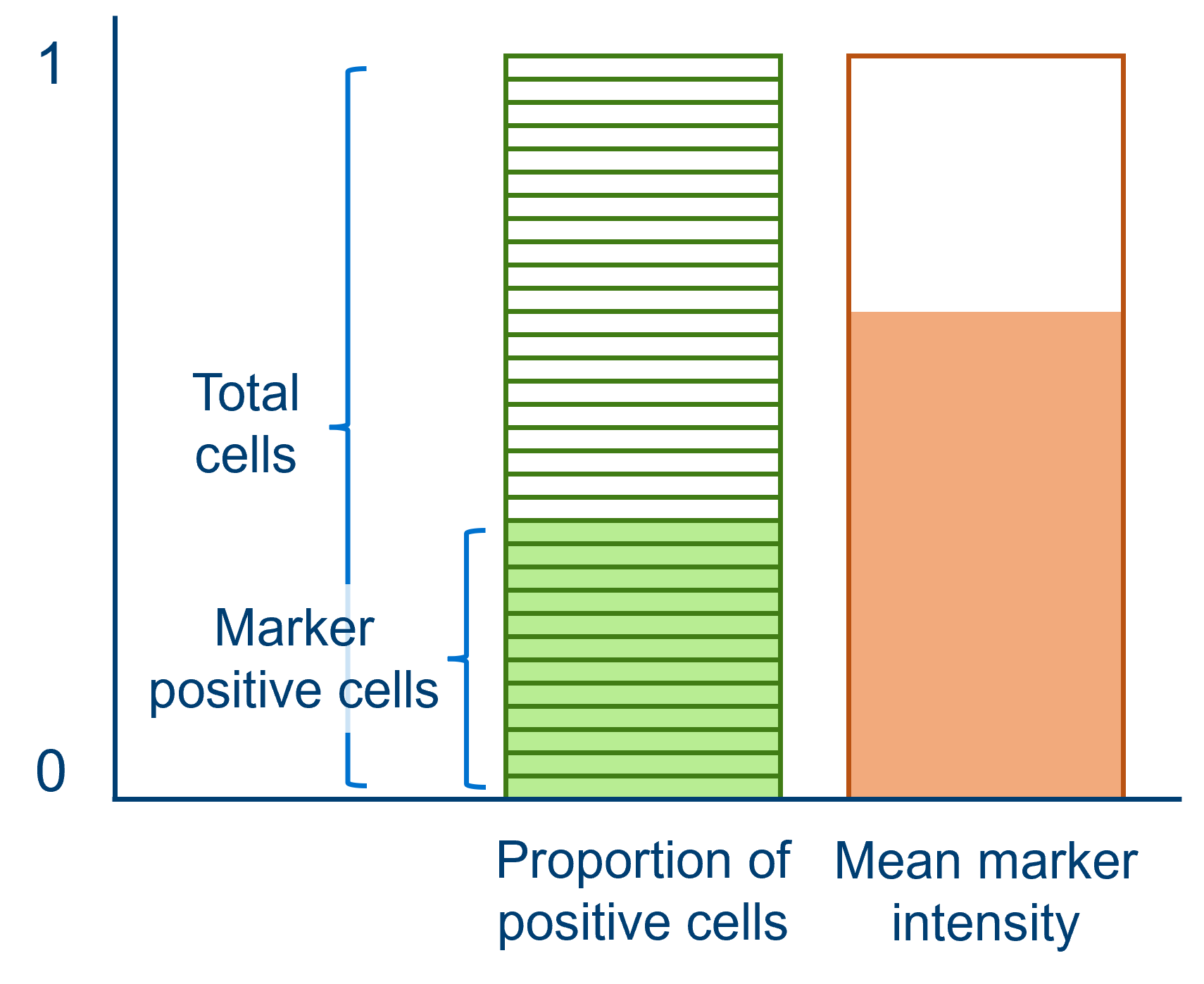
8.7.2 Stem cells
8.8 Summary
- We can use a logistic model for proportional response variables, just as we did with binary variables
- Proportional response variables are made up of a number of trials (success/failure), and should not be confused with fractions/percentages
- Using the equation of a logistic model, we can predict the expected proportion of “successful” trials